- Title
-
Bioinformatic Analysis of Roquin Family Reveals Their Potential Role in Immune System
- Authors
- Li, X., Yang, S., Zhang, X., Zhang, Y., Zhang, Y., Li, H.
- Source
- Full text @ Int. J. Mol. Sci.
|
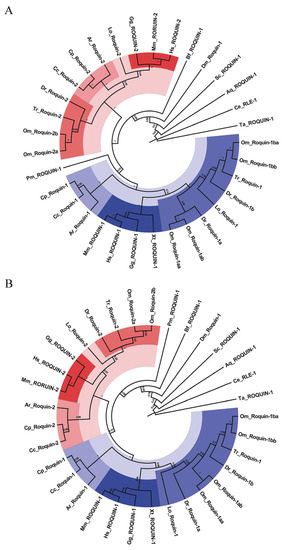
The phylogenetic trees of Roquins between various species constructed by MEGA with two different methods: ( |
|
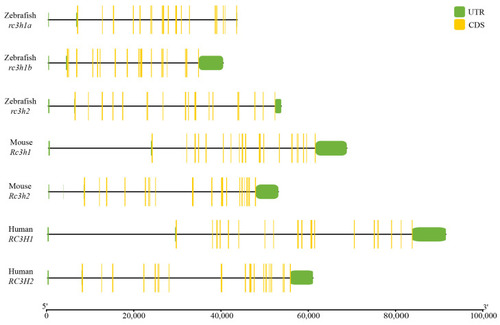
The exon–intron structure of the |
|
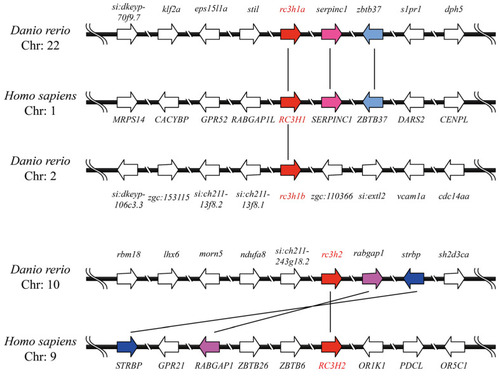
The collinearity analysis of the |
|
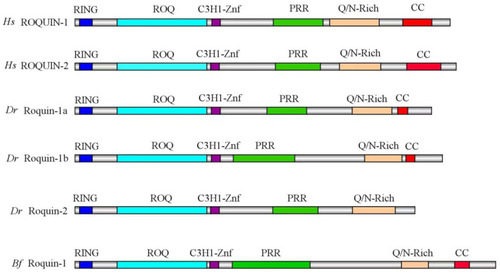
The schematic diagrams of the domain composition of ROQUIN-1 and ROQUIN-2 proteins between various species. RING, ROQ, C3H1-Znf, PRR, Q/N-Rich, and CC domains are shown in colored boxes on a grey background, representing the full length of the proteins. Each domain is depicted with a unique color for visualization. Hs, Homo sapiens; Dr, Danio rerio; Bf, Branchiostoma floridae. |
|
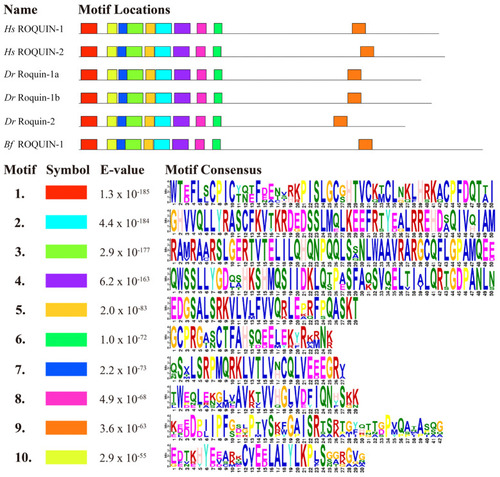
Analysis of the protein motifs of ROQUIN-1 and ROQUIN-2 between various species. The motifs are illustrated in colored boxes. The letters within each motif stand for the abbreviations of amino acids. Larger letters signify higher conservation, indicating a greater probability of the amino acid appearing at the same position within the motif between various species. Hs, Homo sapiens; Dr, Danio rerio; Bf, Branchiostoma floridae. |
|
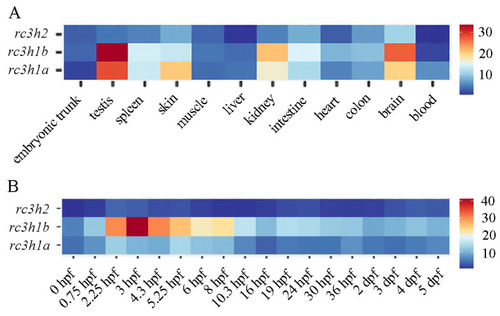
The heatmap displays the expression levels of zebrafish |
|
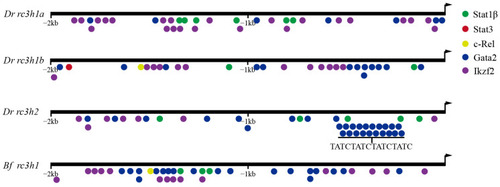
Analysis of transcription factor binding sites in the promoter regions of the |
|
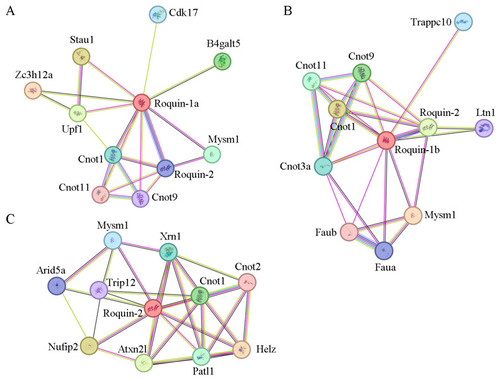
Protein–protein interaction network analysis of Roquin-1a, Roquin-1b, and Roquin-2 in zebrafish. ( |
|
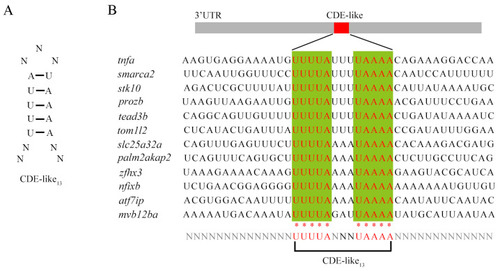
The schematic diagram shows the 3’-UTR region of multiple genes containing CDE-like motifs. ( |