- Title
-
Mapping the Metabolic Characteristics and Perturbation of Adult Casper Zebrafish by Ambient Mass Spectrometry Imaging
- Authors
- Zhou, Z., Sun, Y., Yang, J., Abliz, Z.
- Source
- Full text @ Metabolites
|
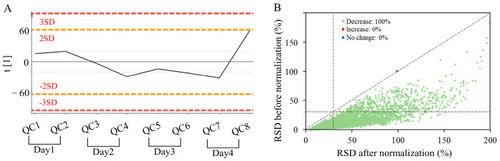
Results of data quality control and normalization obtained by (+) AFADESI-MSI. ( |
|
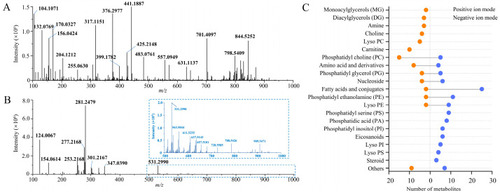
Spatial metabolome of whole-body casper zebrafish obtained by AFADESI-MSI. ( |
|
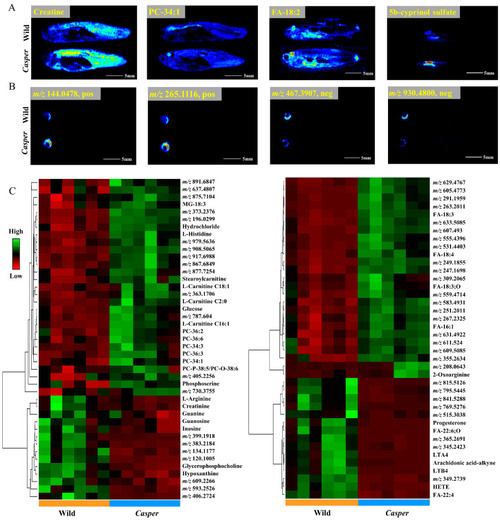
Spatial metabolome differentials between |
|
AFADESI-MSI comparison and statistical analysis results of differential metabolites in altered pathways. ( |
|
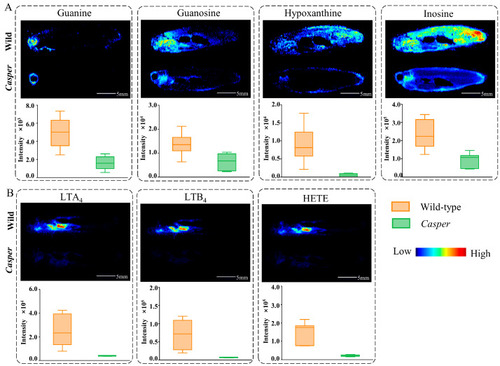
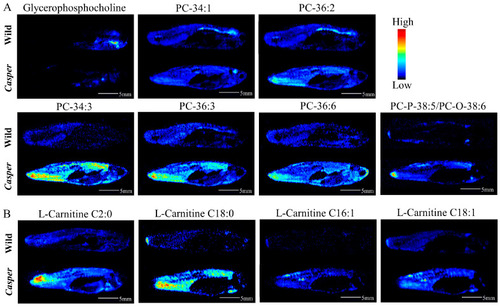
AFADESI-MSI comparison of differential metabolites between |
|
Metabolic disorder in |