- Title
-
The Peptide LLTRAGL Derived from Rapana venosa Exerts Protective Effect against Inflammatory Bowel Disease in Zebrafish Model by Regulating Multi-Pathways
- Authors
- Cao, Y., Xu, F., Xia, Q., Liu, K., Lin, H., Zhang, S., Zhang, Y.
- Source
- Full text @ Mar. Drugs
|
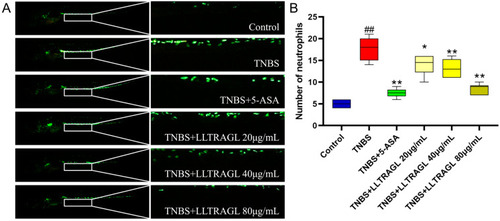
Effect of peptide LLTRAGL on TNBS-induced migration of zebrafish to intestinal macrophages. ( |
|
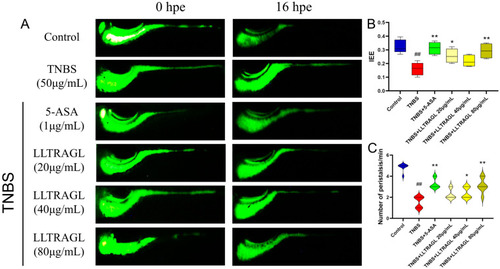
LLTRAGL improves TNBS-induced intestinal peristalsis damage. ( |
|
Effects of the peptide on TNBS-induced intestinal tissue pathology and ultrastructure of zebrafish. ( |
|
The differentially expressed genes among the control group, TNBS group and peptide treatment group in RNA-Seq. ( |
|
GO enrichment analysis of DEGs and GSEA of all tested genes. ( |
|
KEGG pathway enrichment analysis of DEGs. ( |
|
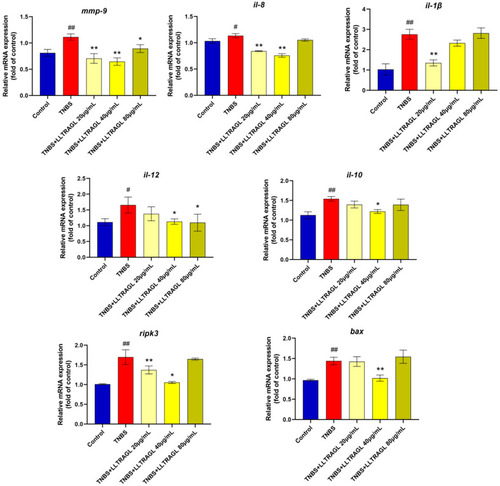
Effect of LLTRAG on the inflammation-related gene expression in zebrafish, determined by RT-PCR. Compared with the blank group, # |
|
Effect of LLTRAG on gene expression in TNBS-induced colitis in zebrafish. ( |