- Title
-
Replacement of Dietary Fish Protein with Bacterial Protein Results in Decreased Adiposity Coupled with Liver Gene Expression Changes in Female Danio rerio
- Authors
- Williams, M.B., Green, G.B., Palmer, J.W., Fay, C.X., Chehade, S.B., Lawrence, A.L., Barry, R.J., Powell, M.L., Harris, M.L., Watts, S.A.
- Source
- Full text @ Curr Dev Nutr
|
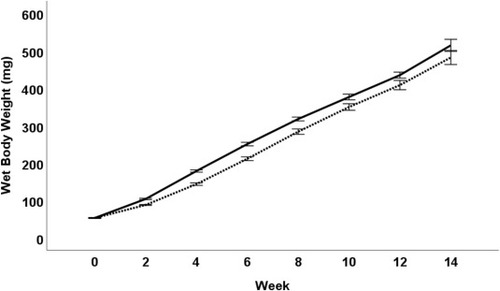
Total body weight average for tanks of fish (mg) for male and female zebrafish (combined) fed either BP (black line) or SR (dashed line) protein diets measured every 2 wk from week 2 to week 14 on the assigned diets (n = 10 tanks, 14 fish per tank for each diet treatment). P value represents linear regression of the trend line. BP, bacterial protein; SR, standard reference. |
|
Terminal wet body weight (mg; A), standard length (mm; B), total body moisture (%; C), and dry body lipid (%; D) averages for individual fish for male and female zebrafish (separated) measured at termination at 14 wk on the assigned diets (n = 10 tanks, 14 fish per tank for each diet treatment). |
|
Total egg produced (A), egg viability at 4 hpf (B), and egg viability at 24 hpf (C) averages for breeding of female zebrafish after 14–18 wk on the assigned diets crossed with males fed Artemia (n = 20 breeding events for female of the dietary treatments with a 2-wk gap). |
|
Principal component analysis (PCA) among samples, red dots represent the female D. rerio fed with the fish protein hydrolysate diet [standard reference (SR)], and the blue dots represent the female D. rerio fed with the single-cell bacterial protein (BP) diet. The PCA plot was generated via DeSeq2 (ver. 1.22.2). |
|
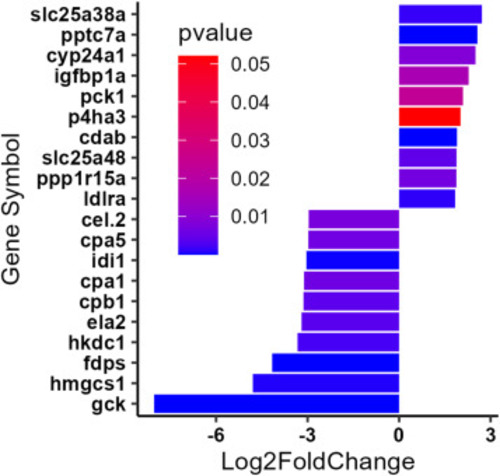
The top 10 upregulated and top 10 downregulated differentially expressed genes (DEGs). The genes were selected after differential expression analysis (DeSeq2), and were filtered based on a BaseMean > 500, log2FC > |1.5| and a P value < 0.1. |
|
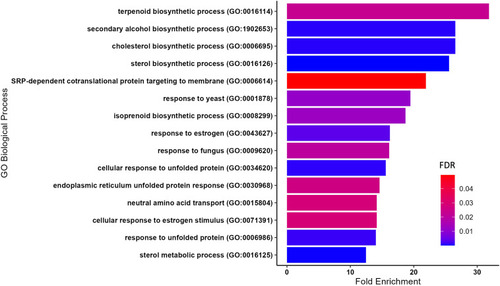
Top 15 most significantly enriched gene ontologies (Gene Ontology, released 3 January, 2023) based on the highest fold enrichment, and a FDR < 0.05, were plotted in R, using ggplot2 (v3.4.0). FDR, false discovery rate. |
|
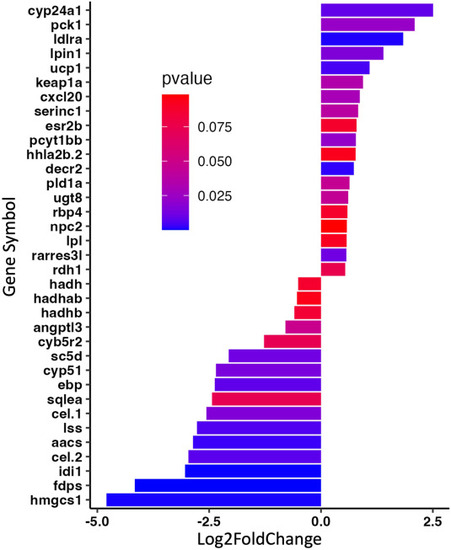
Genes associated with lipid-related pathways determined via GO analysis. These pathways were: “sterol biosynthetic bioprocess,” “response to lipid,” “fatty metabolic process,” sterol metabolic process,” “lipid metabolic process,” “fatty acid catabolic process,” and “lipid biosynthetic process.” Genes with a log2foldchange > |1.5|, and a P value < 0.1 were plotted via a ggplot2 (v.3.4.0). GO, gene ontology. |
|
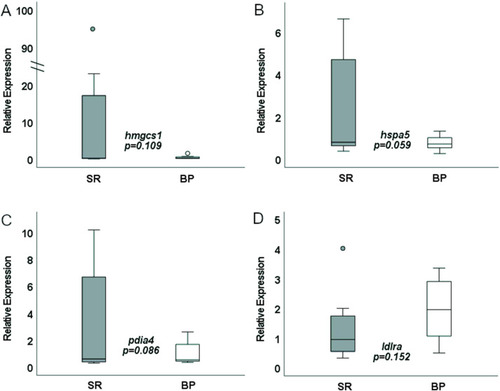
rtPCR liver expression of SR- and BP-fed females (n = 8 livers). BP, bacterial protein; SR, standard reference. |
|
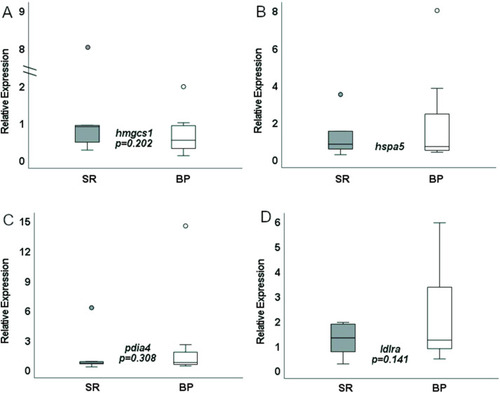
rtPCR liver expression of SR- and BP-fed males (n = 5–7 livers). BP, bacterial protein; SR, standard reference. |