- Title
-
eIF4E1b is a non-canonical eIF4E protecting maternal dormant mRNAs
- Authors
- Lorenzo-Orts, L., Strobl, M., Steinmetz, B., Leesch, F., Pribitzer, C., Roehsner, J., Schutzbier, M., Dürnberger, G., Pauli, A.
- Source
- Full text @ EMBO Rep.
|
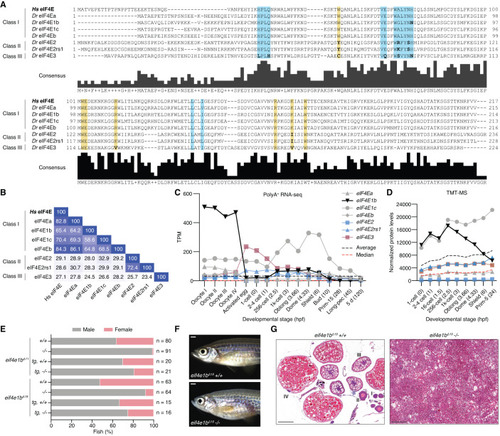
eIF4E1b is a class I eIF4E protein with an essential role in zebrafish oogenesis. ( |
|
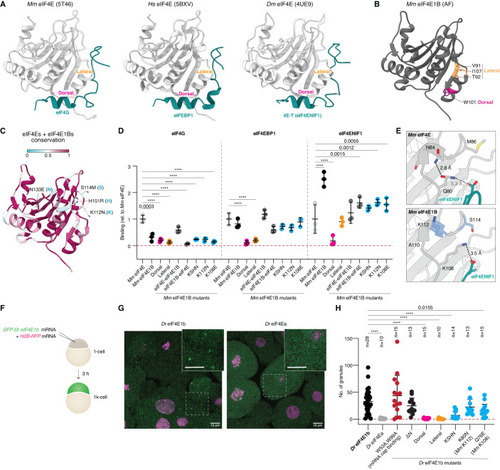
eIF4E1b interacts with the mRNA cap, eIF4EBP1, and eIF4ENIF1 and localizes to cytoplasmic foci in oocytes and embryos. ( |
|
Specific eIF4E1b residues mediate eIF4ENIF1 binding and localization to P-bodies in the embryo. ( |
|
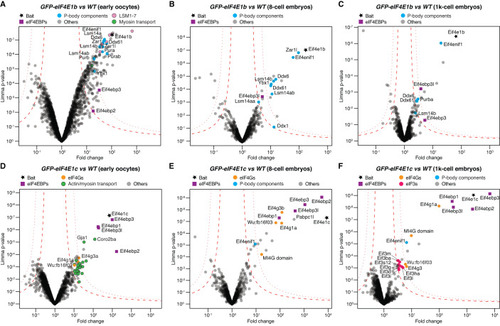
eIF4E1b interacts with P-body components in zebrafish oocytes and embryos. Volcano plots of proteins enriched by immunoprecipitation followed by mass spectrometry (IP-MS) using GFP-tagged eIF4E1b ( |
|
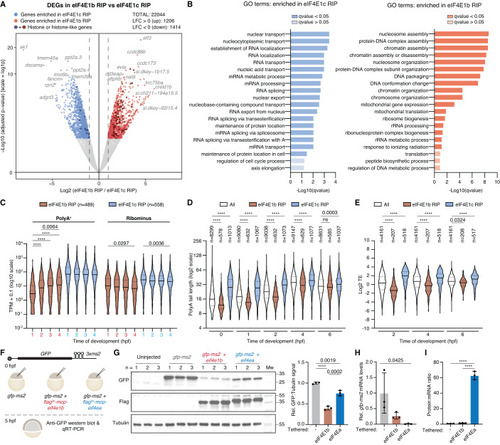
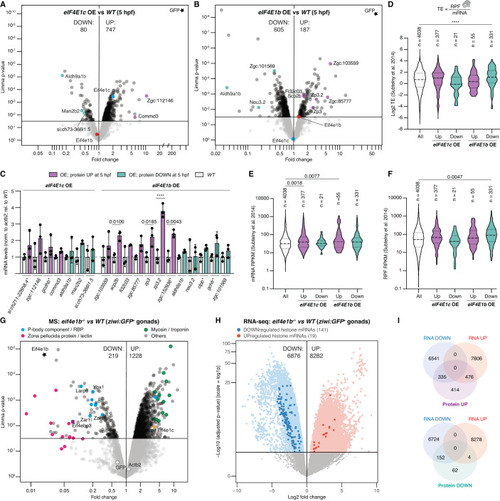
eIF4E1b binds to translationally repressed mRNAs involved in chromatin regulation. ( |
|
eIF4E1b levels negatively correlate with total protein levels in zebrafish embryos and gonads. ( |