- Title
-
Modulation of Zebrafish (Danio rerio) Intestinal Mucosal Barrier Function Fed Different Postbiotics and a Probiotic from Lactobacilli
- Authors
- Rawling, M., Schiavone, M., Mugnier, A., Leclercq, E., Merrifield, D., Foey, A., Apper, E.
- Source
- Full text @ Microorganisms
|
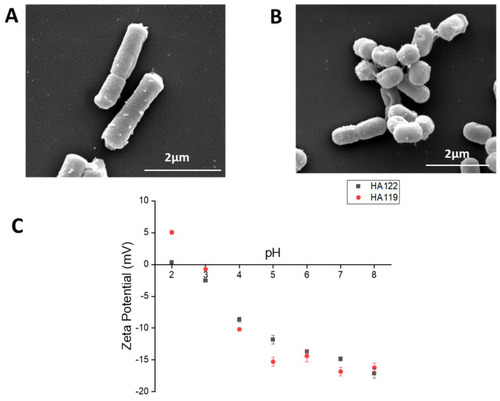
Morphology and surface charge of LHPost and LPPost. ( |
|
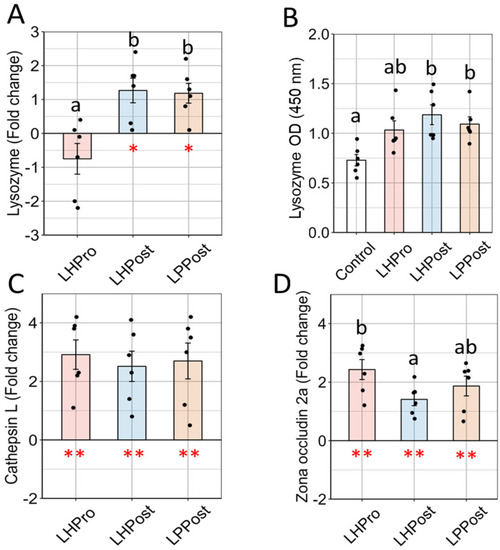
Elevated phenotypic markers showing the fortification of the mucosal barrier in fish fed diets supplemented with postbiotics compared to other treatments. ( |
|
Gene expression analysis revealed positive elevations in markers, suggesting a fortification of the mucosal barrier in fish fed diets supplemented with postbiotics compared to other groups. ( |
|
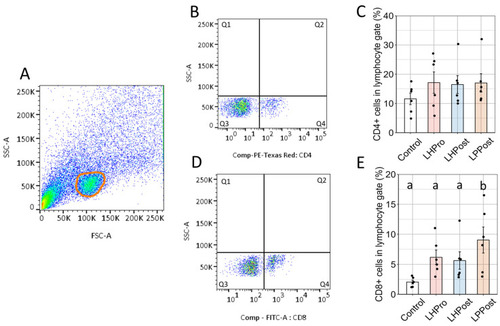
Mobilization of positive CD8α+ cells in the posterior intestine of zebrafish fed probiotic and postbiotic treatments. ( |
|
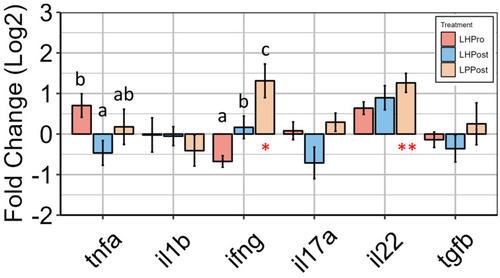
Gene expression analysis reveals the modulation of innate immune effector cytokines by postbiotic groups in the posterior intestine of zebrafish. Relative expression level (fold change (Log2)) to the control group of innate immune markers is shown. Data are presented as mean ± SEM. Symbols in red denote significance in gene expression compared to the control group (* |
|
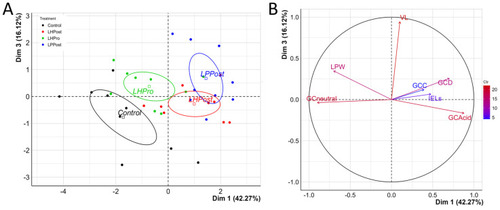
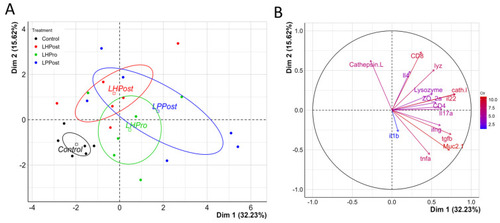
Positive differentiation of immune response markers as shown by principal component analysis. ( |