- Title
-
Genetic model of UBA5 deficiency highlights the involvement of both peripheral and central nervous systems and identifies widespread mitochondrial abnormalities
- Authors
- Serrano, R.J., Oorschot, V., Palipana, D., Calcinotto, V., Sonntag, C., Ramm, G., Bryson-Richardson, R.J.
- Source
- Full text @ Brain Commun
|
|
|
|
|
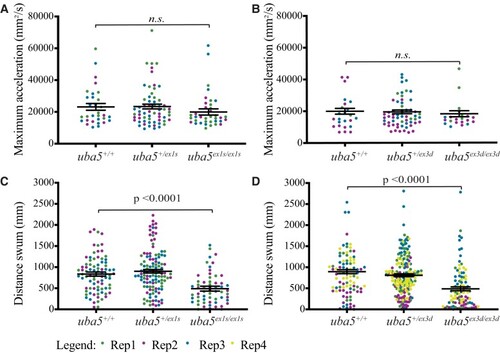
PHENOTYPE:
|
|
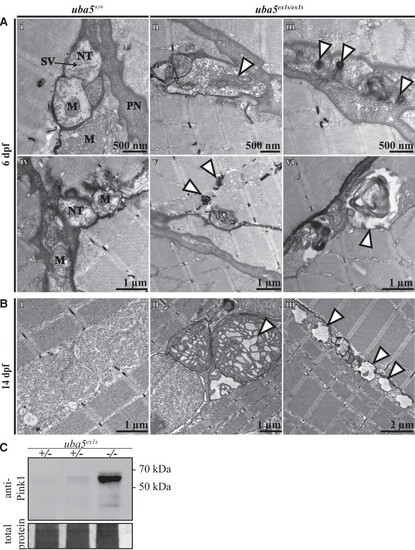
EXPRESSION / LABELING:
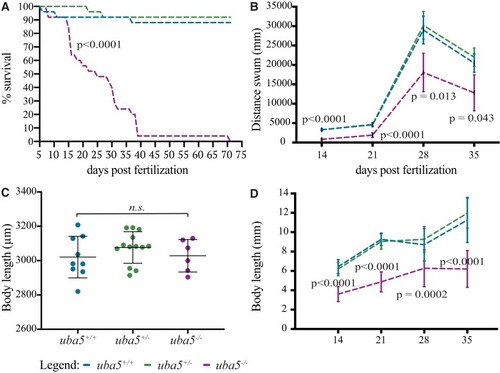
PHENOTYPE:
|
|
|
|
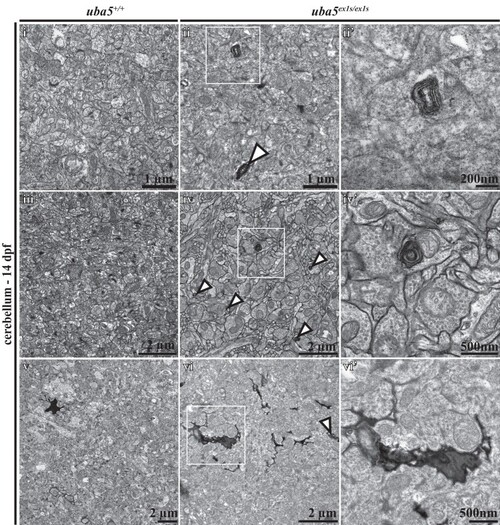
PHENOTYPE:
|