- Title
-
asb5a/asb5b Double Knockout Affects Zebrafish Cardiac Contractile Function
- Authors
- Cai, W., Wang, Y., Luo, Y., Gao, L., Zhang, J., Jiang, Z., Fan, X., Li, F., Xie, Y., Wu, X., Li, Y., Yuan, W.
- Source
- Full text @ Int. J. Mol. Sci.
|
Schematic diagrams of |
|
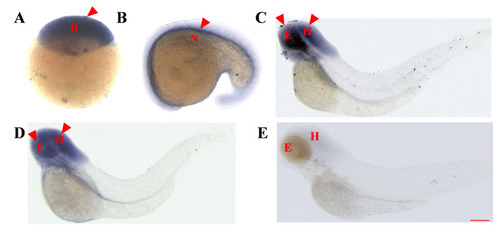
Temporal and spatial expression patterns of the |
|
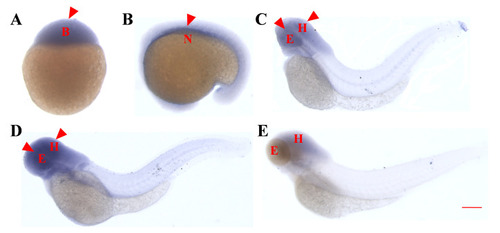
Temporal and spatial expression pattern of the |
|
Effects of |
|
Effects of |
|
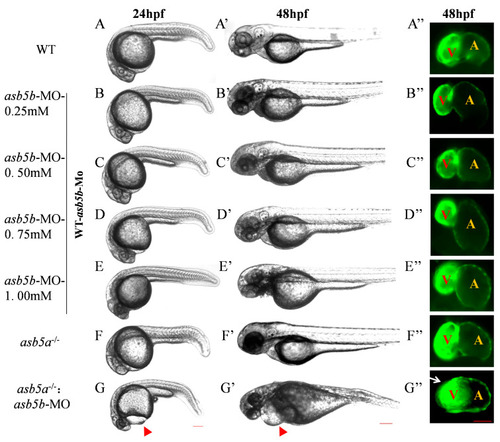
Effects of |
|
Effect of |
|
M-mode revealed cardiac physiological functions after |
|
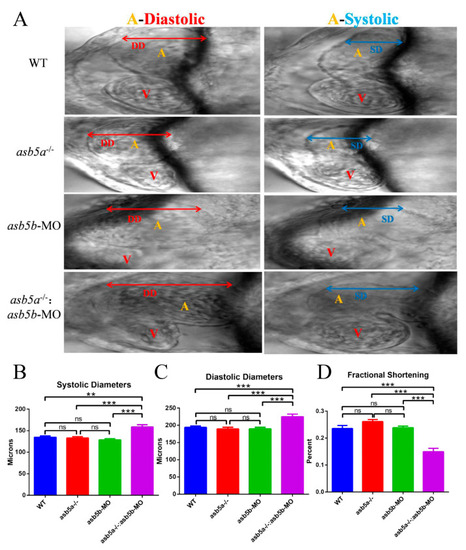
Atrial morphological changes after |
|
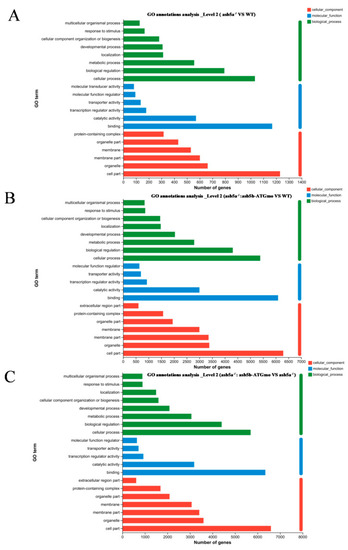
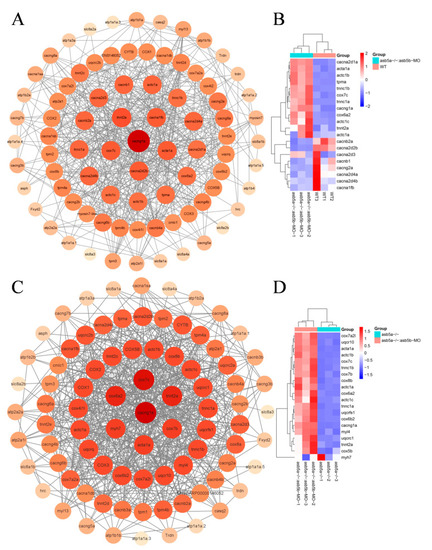
PPI network analysis and heatmap of the top 20 hub genes. ( |
|
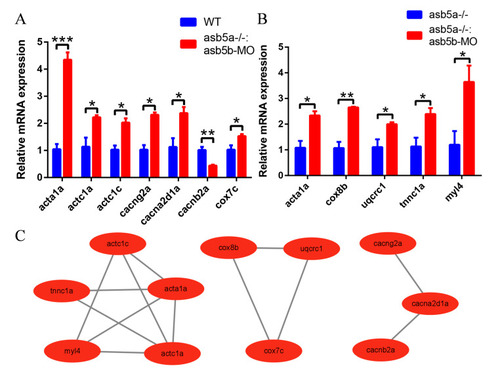
qPCR validation and the PPI network diagram. ( |
|
Probable molecular regulatory mechanism of 11 hub genes affecting cardiac contractile function following |