- Title
-
Endogenous tenocyte activation underlies the regenerative capacity of the adult zebrafish tendon
- Authors
- Tsai, S.L., Villaseñor, S., Shah, R.R., Galloway, J.L.
- Source
- Full text @ NPJ Regen Med
|
The adult zebrafish can fully regenerate after acute injury. |
|
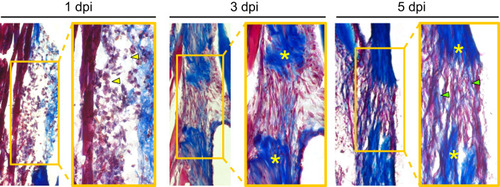
Tendon regeneration proceeds through a rapid series of phases within the first week post-injury. Masson’s trichrome staining of sections from regenerating tendons at 1 (left), 3 (middle), and 5 (right) dpi. Heavy infiltration of cells with myeloid-like morphologies can be seen at 1 dpi (yellow arrowheads). At 3 dpi, a fibroblastic bridge connecting the two severed tendon ends is evident. By as early as 5 dpi, the beginnings of collagen matrix deposition into the injury site are observed (green arrowheads). Yellow asterisks denote severed tendon ends and images were taken at 10x magnification. Dpi, days post-injury. |
|
Tendon injury triggers a rapid innate immune response followed by a wave of cellular proliferation. |
|
Generation and validation of a |
|
Pre-existing tenocytes are a major cell source of tendon regeneration. |
|
TGF-β signaling is active in tenocytes during regeneration. |
|
Canonical TGF-β signaling is required for adult zebrafish tendon regeneration. |
|
TGF-β signaling is required for tenocyte recruitment, but not proliferation, during bridge formation. |
|
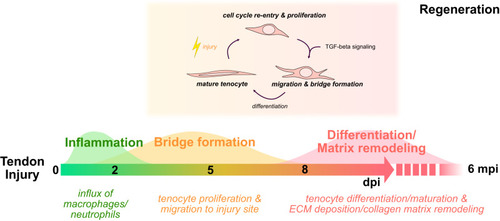
Hallmarks of adult zebrafish tendon regeneration. Schematic detailing the timeline of key cellular processes following acute tendon injury and regeneration as well as the requirement of TGF-β signaling for tenocyte recruitment. dpi days post-injury, mpi months post-injury. |