- Title
-
The Presence of Two MyoD Genes in a Subset of Acanthopterygii Fish Is Associated with a Polyserine Insert in MyoD1
- Authors
- White, L.J., Russell, A.J., Pizzey, A.R., Dasmahapatra, K.K., Pownall, M.E.
- Source
- Full text @ J Dev Biol
|
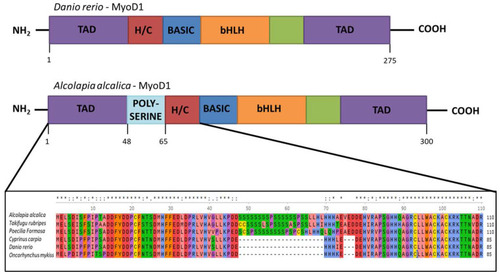
Inclusion of a polyserine insertion in MyoD1 in some fish species. Comparison of the protein structure of MyoD1 from Danio rerio (above) and Oreochromis (Alcolapia) alcalica (below). The enlargement of the schematic shows an alignment of the amino acid sequences from the indicated part of MyoD1 proteins from Oreochromis (Alcolapia) alcalica, Takifugu ruberipes, Poecilia formosa, Cyprinus carpio, Danio rerio, Oncorhynchus mykiss including the polyserine insertion in some species. Multiple species alignment of MyoD1 with and without the polyserine insertion. * represents exact match and: represents residues where the majority of species show conservation. |
|
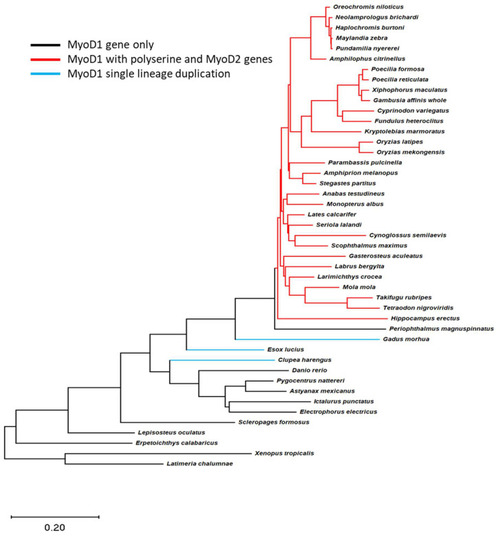
A polyserine insert is found in MyoD1 proteins from species that retain both MyoD1 and MyoD2 genes. Teleost species phylogeny is shown for all species with complete MyoD1 and MyoD2 data available; the tree by [20] was trimmed using R packages to only include those species with data available. Gene information was subsequently overlaid to this tree and branches coloured to represent the species differences in MyoD genes found in the genome. Black lines indicate species with a single MyoD gene with no polyserine insert. Red lines indicate the presence of MyoD1 and MyoD2 genes where the MyoD1 sequence includes a polyserine insertion. The blue line indicates a lineage specific duplication of MyoD1; MyoD1 proteins in this case do not include a polyserine insert. The phylogeny indicates that the polyserine insertion in MyoD1 is only found in species which have retained a MyoD2 gene. |
|
Expression analysis of MyoD1 and MyoD2 in the developing embryos of Oreochromis (Alcolapia) alcalica. (A,C,E) Lateral and dorsal views of O. alcalica embryos analysed by in situ hybridisation for MyoD1 and (B,D,F) for MyoD2. The number of days post fertilisation [dpf] is indicated for each image. Arrows show somites, white arrowheads indicate facial muscle (FM) and white arrows indicate developing pectoral fin bud (PFB). Black dots around the yolk and on the body are chromatophores (pigment cells). |
|
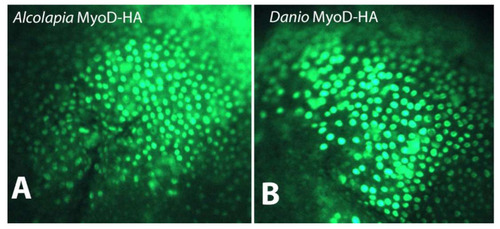
Subcellular localisation of MyoD1 proteins. Immunostaining for HA-tagged MyoD1 proteins from (A) O. alcalica and (B) D. rerio shows nuclear localisation when mRNAs coding for these proteins are expressed in Xenopus laevis embryos. |
|
Stability assay shows more perdurance of O.alcalica MyoD1 compared to D. rerio MyoD1. (A) Western blot analysis of extracts from Xenopus laevis explants overexpressing HA-tagged MyoD1 proteins. Explants were incubated for either 0 (T0), 1 (T1) or 3 h (T2) in the translational inhibitor, cycloheximide. Anti- beta tubulin is a loading control. (B) Quantification of the loss of protein relative to the amount of protein detected at T0 is shown in a graph for O.alcalica MyoD1 and for D. rerio MyoD1. |
|
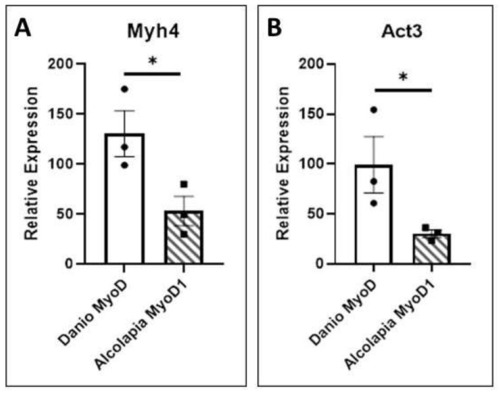
O. alcalica MyoD1 and D. rerio MyoD1 activate muscle gene expression. (A) The expression of myosin 4 (myh4) is quantified by qRT-PCR analysis of RNA extracted from explants expressing O.alcalica MyoD1 compared to D. rerio MyoD1. (B) The expression of actin 3 (act3) is quantified by qRT-PCR analysis of RNA extracted from explants expressing O.alcalica MyoD1 compared to D. rerio MyoD1. While both MyoD1 proteins can activate high levels of contractile gene expression in naive tissue, there is a significantly higher response to D. rerio MyoD1. Experiment was carried out in triplicate and average Ct was normalised to the housekeeping gene Dicer. Pairwise t-tests comparing the mean relative expression for control and experimental sets for each gene. Error bars represent SEM, * = p < 0.05. |