- Title
-
Investigating the effects of chronic low-dose radiation exposure in the liver of a hypothermic zebrafish model
- Authors
- Cahill, T., da Silveira, W.A., Renaud, L., Wang, H., Williamson, T., Chung, D., Chan, S., Overton, I., Hardiman, G.
- Source
- Full text @ Sci. Rep.
|
( |
|
( |
|
( |
|
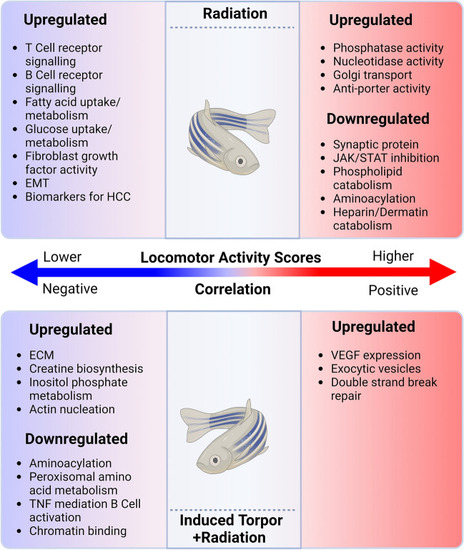
Genes correlated with locomotor activity scores in the radiation and torpor + radiation groups. This schematic shows that increased activity scores in the radiation group are correlated to an upregulation of genes involved in biological processes such as nucelotidase activity, Golgi transport and phagocytosis, and a downregulation of growth, endocytosis, aminoacylation and heparin catabolism related genes. Genes correlated with lower activity scores in the group are associated with immune-related processes such as hemopoiesis, mononuclear cell differentiation, and B cells receptor signalling. In the torpor + radiation group genes correlated with higher activity scores are associated with VEGF expression, exocytic vesicles, and double strand break repair. Genes correlated with decreased activity are correlated with ECM, actin nucleation, creatine biosynthesis and inositol phosphate and a downregulation of genes involved in aminoacylation, amino acid metabolism, B cell receptor activation and chromatin binding. Created with BioRender.com. |
|
( |
|
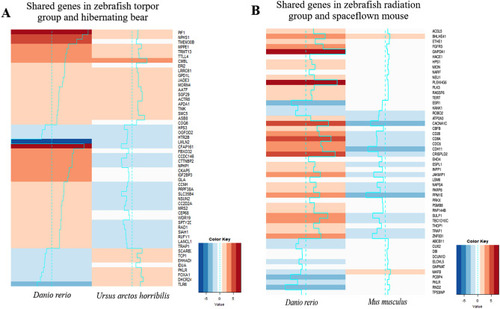
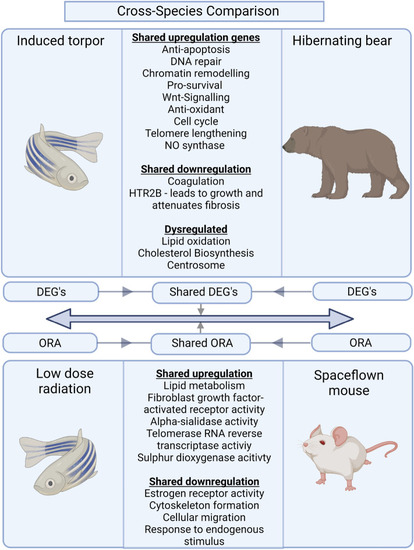
Comparison of our zebrafish radiation group vs space flown mouse and the zebrafish induced torpor group vs hibernating bear. The results between the zebrafish radiation model and the space flown mouse show shared upregulation of genes involved in sulphur dioxygenase activity, telomerase RNA reverse transcriptase, alpha-sialidase activity, and fibroblast growth factor receptor activity. Shared downregulated genes included the estrogen receptor ( |