- Title
-
The Annotation of Zebrafish Enhancer Trap Lines Generated with PB Transposon
- Authors
- Jia, W., Guan, Z., Shi, S., Xiang, K., Chen, P., Tan, F., Ullah, N., Diaby, M., Guo, M., Song, C., Gao, B.
- Source
- Full text @ Curr. Iss. Mol. Biol.
|
GFP expression patterns in the EYE line generated by the PB-mediated enhancer trap. (A?I) nine developmental stages. The arrows indicate GFP+ tissues. |
|
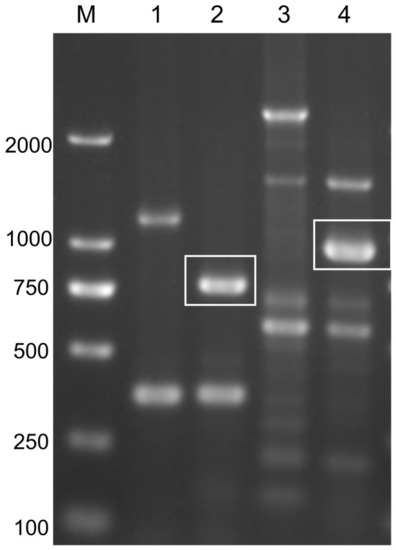
Electrophoresis image of PCR products in agarose gel. M: DL2000 bp marker; Line 1: the PCR product amplified from wild-type zebrafish genome by using GFP-F/R primers; Line 2: the PCR product amplified from EYE lines genome by using GFP-F/R primers; Line 3: the PCR product amplified from wild-type zebrafish genome by using Krt4-F and GFP-R primers; 4: the PCR product amplified from EYE lines genome by using Krt4-F and GFP-R primers. White boxes indicate the target bands (720 bp in Line 2, 916 bp in Line 4). |
|
Enhancer distribution between 50 kb upstream and 50 kb downstream of the insertion site in the EYE lines genome. Genomic sequences from Danio rerio, Northern-pike, Nile-tilapia, Mummichog, Midas-cichlid, Mexican tetra, and Amazon-molly were analyzed in the VISTA browser. The red boxes represent three conserved non-coding sequences (CNS) that are predicted to be enhancers between itgav and zc3h15 genes. The red arrow indicates the insertion site of the ET vector. UTR: untranslated region. |
|
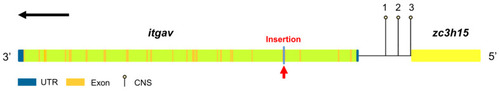
Itgav structural map. The red arrow indicates the insertion site of the ET vector. The black arrow indicates the orientation of the gene itgav. Numbers 1, 2, 3 indicate three conserved non-coding sequences (CNS) located upstream of itgav or downstream of zc3h15. UTR: untranslated region; CNS: conserved non-coding sequence. |
|
In situ hybridization for itgav transcripts at different stages of wide type zebrafish embryos. Red arrows indicate similar expression patterns to GFP in the EYE line. Experimental group: zebrafish embryos treated with itgav antisense RNA probes. Control group: zebrafish embryos without probe treatment. |