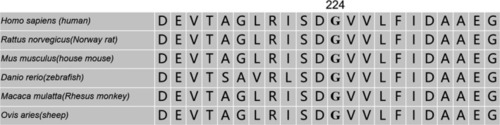- Title
-
A novel de novo missense mutation in EFTUD2 identified by whole-exome sequencing in mandibulofacial dysostosis with microcephaly
- Authors
- Yang, M., Liu, Y., Lin, Z., Sun, H., Hu, T.
- Source
- Full text @ J Clin Lab Anal
|
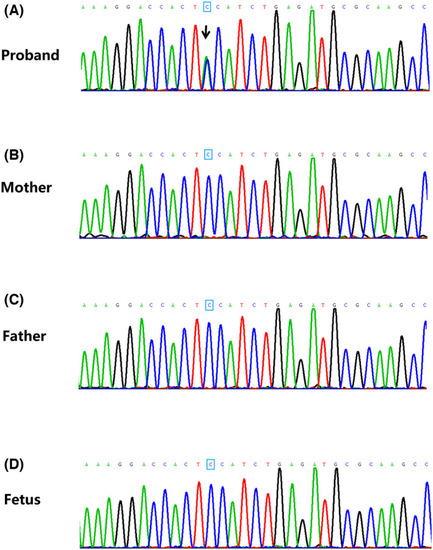
Sanger sequencing of the novel EFTUD2 missense mutation. Whole‐exome sequencing identified a novel EFTUD2 missense mutation (c.671G>T, p.Gly224Val) in a proband with mandibulofacial dysostosis with microcephaly (MFDM). (A) Sanger sequencing confirmed the EFTUD2 missense mutation (c.671G>T, p.Gly224Val) in blood samples from the proband; (B‐D) the EFTUD2 missense mutation (c.671G>T, p.Gly224Val) was not detected in peripheral blood samples of the proband's parents and amniotic fluid samples of the parents’ second baby. Black arrows indicate the point mutation (G>T) |
|
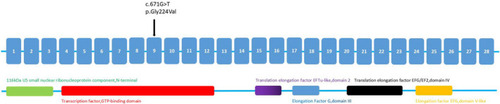
Protein domains and location of mutation in EFTUD2. The missense mutation (c.671G>T) in EFTUD2 was located within the transcription factor GTP‐binding domain, and prior to translation elongation factor EFTu‐like, domain 2. Black arrows indicate the point mutation (G>T) |
|
Gly224 is highly conserved in many vertebrate species |
|
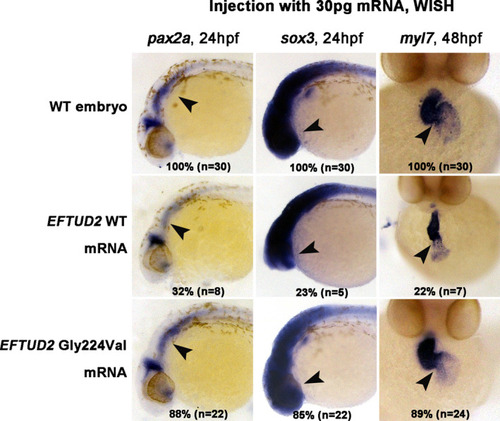
p.Gly224Val mutation leads to loss of function of EFTUD2 in the zebrafish model. p.Gly224Val mutation caused a loss of gene function of EFTUD2 during zebrafish embryogenesis. The EFTUD2 p.Gly224Val mutant mRNA decreased percentage of abnormality of embryonic neurodevelopment and cardiac development compared to EFTUD2 WT mRNA. Expression of pax2a and sox3 at 24 hpf, myl7 at 48 hpf, embryos at 24 hpf shown are lateral views with anterior to the left, embryos at 48 hpf shown are ventral view with the anterior at the top. Arrows point to the signal generated by detected marker genes. The percentage and numbers indicated in each image are the ratio for the number of affected embryos with phenotype similar to what is shown in the picture |