- Title
-
Hearing loss genes reveal patterns of adaptive evolution at the coding and non-coding levels in mammals
- Authors
- Trigila, A.P., Pisciottano, F., Franchini, L.F.
- Source
- Full text @ BMC Biol.
|
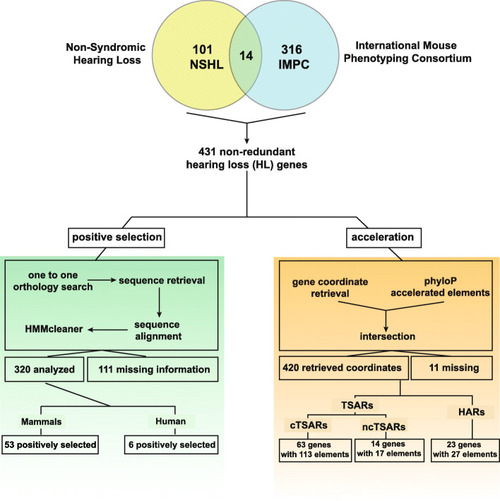
Bioinformatics pipeline and filtering process of two datasets of hearing loss genes: the classic dataset composed of non-syndromic hearing loss genes identified in humans which is composed of a total of 129 coding genes obtained by combining the information available in the “Hereditary Hearing Loss Homepage” |
|
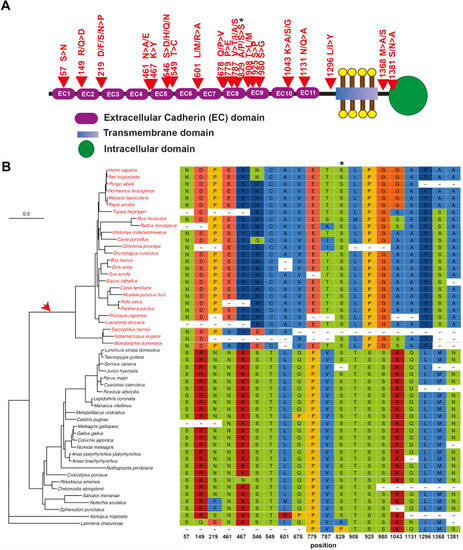
Phylogenetic tree and positive selected sites of an essential tip link protein: PCDH15. |
|
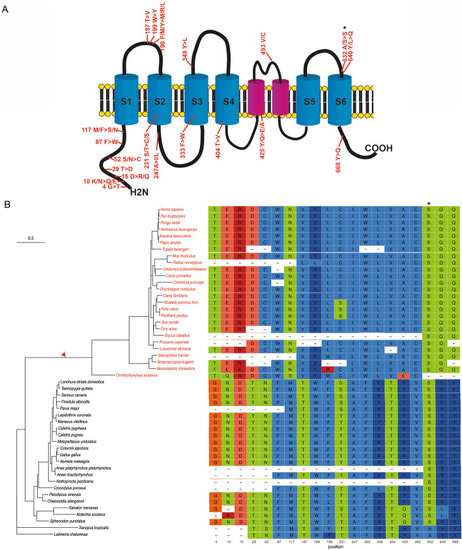
Phylogenetic tree and positive selected sites of an essential tip link protein: TMC1. |
|
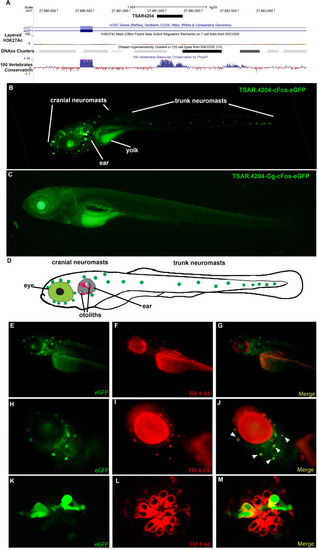
Representative transgenic zebrafish carrying the TSAR4204-JAZF1. A Location of the TSAR.4204 in the JAZF1 locus in chromosome 7 of the human genome (modified from UCSC Genome Browser). B Low-magnification microphotograph showing the expression pattern of the transgene TSAR.4204-cFos-eGFP (human sequence as a mammalian representative) in a transgenic zebrafish at 6 dpf. C Low-magnification microphotograph showing the expression pattern of the transgene TSAR.4204-Gg-cFos-eGFP carrying the chicken ortholog of the TSAR.4204 sequence in a transgenic zebrafish at 6 dpf. D Scheme of a 6 dpf zebrafish showing the approximate location of the inner ear and neuromasts of the lateral line, where the transgene carrying the mammalian sequence of TSAR.4204 directs the expression of eGFP. Figures (E–M) show magnifications of the head region of a transgenic zebrafish carrying the human sequence of TSAR.4204-cFos-eGFP exhibiting in major detail the expression of eGFP (E, H, K), the fluorescent marker FM4-64 that specifically labels hair cells (F, I, L), and a merged image showing the colocalization of eGFP and the hair cell marker (G, J, M). Figures (K–M) show in detail confocal images of neuromast expressing eGFP in the lateral line of transgenic zebrafish carrying the human sequence of TSAR.4204-cFos-eGFP. We show the best representative image for each line |
|
Key genes involved in the mechanotransduction machinery display signatures of positive selection. |