- Title
-
Pigment pattern morphospace of Danio fishes: evolutionary diversification and mutational effects
- Authors
- McCluskey, B.M., Liang, Y., Lewis, V.M., Patterson, L.B., Parichy, D.M.
- Source
- Full text @ Biol. Open
|
|
|
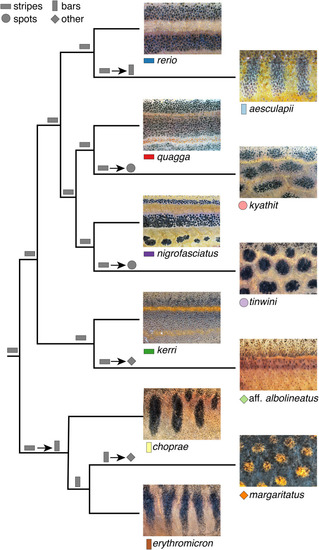
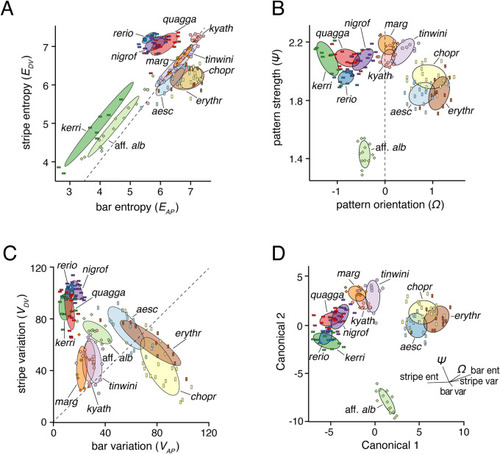
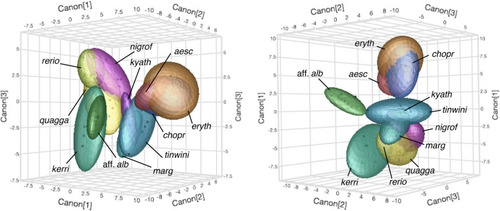
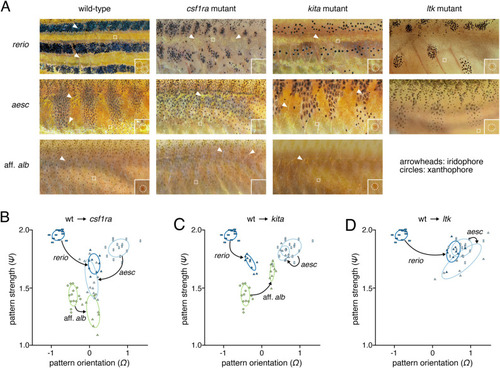
Species pattern variation. Panels show typical wild-type phenotypes within species and illustrate the regions of interest and greyscale conversions used for analyses. Left two columns show females, right two columns show males. |
|
|
|
|
|
|
|
|