- Title
-
Identification and characterization of amphibian SLC26A5 using RNA-Seq
- Authors
- Wang, Z., Wang, Q., Wu, H., Huang, Z.
- Source
- Full text @ BMC Genomics
|
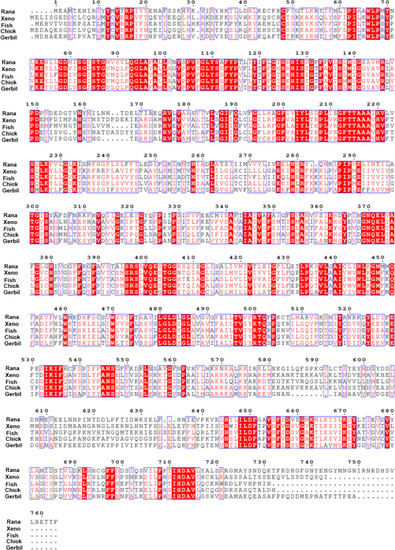
Alignment of amino acid sequences of rana (American bullfrog), xeno (tropical clawed frog), fish (zebrafish), chick and gerbil prestin. Different colors had been used to represent identity of each residue among two species. Red block: Full identity at a residue; red letter: Partial identity at a residue; Black: complete disparity at a residue. Gaps in the aligned sequences were indicated by the dashed line |
|
Frog prestin localized in the cell membrane. |
|
The nonlinear capacitance measurements in the hair cells from bullfrog’s amphibian papilla (AP) and in the exogenous expressed cells. |
|
NLC datas of rana’s amphibian papilla (AP) hair cells and rana prestin transfected cells (rPres). |
|
NLC measurements from rana, xenopus and chick. |
|
Comparing the NLC datas of xenopus prestin with rana prestin. |