- Title
-
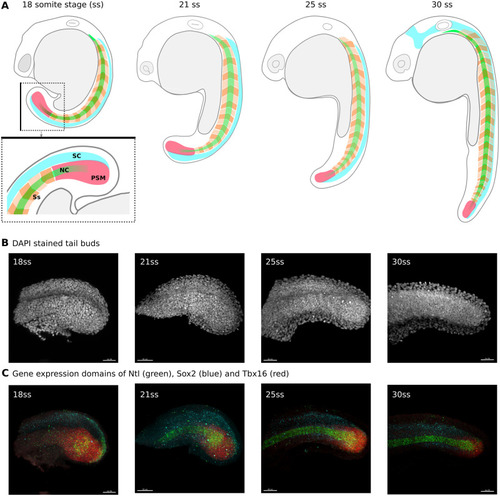
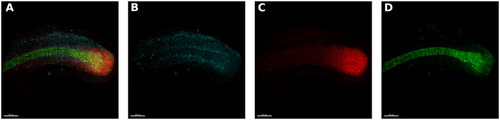
A deep learning approach for staging embryonic tissue isolates with small data
- Authors
- Pond, A.J.R., Hwang, S., Verd, B., Steventon, B.
- Source
- Full text @ PLoS One
|
|
|
|
|
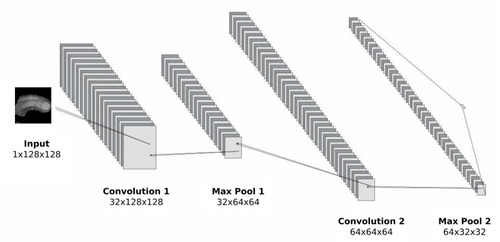
Convolution and pooling region of the 2D CNN architecture. |