- Title
-
The Genome-Wide Impact of Nipblb Loss-of-Function on Zebrafish Gene Expression
- Authors
- Spreafico, M., Mangano, E., Mazzola, M., Consolandi, C., Bordoni, R., Battaglia, C., Bicciato, S., Marozzi, A., Pistocchi, A.
- Source
- Full text @ Int. J. Mol. Sci.
|
( |
|
( |
|
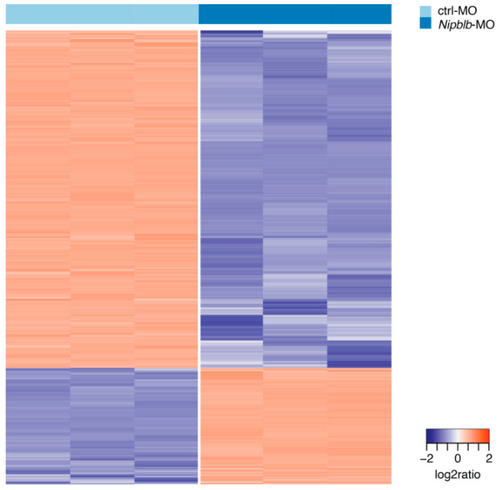
Hierarchical clustering of ctrl-MO and |
|
( |
|
( |