- Title
-
Anti-inflammatory and Regulatory Effects of Huanglian Jiedu Decoction on Lipid Homeostasis and the TLR4/MyD88 Signaling Pathway in LPS-Induced Zebrafish
- Authors
- Zhou, J., Gu, X., Fan, X., Zhou, Y., Wang, H., Si, N., Yang, J., Bian, B., Zhao, H.
- Source
- Full text @ Front. Physiol.
|
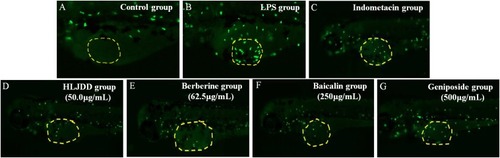
Representative pictures of transgenic neutrophils labeling in zebrafish yolks at 3 dpf. |
|
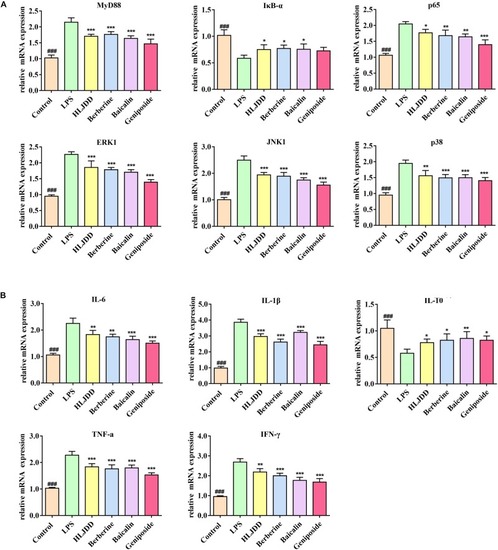
Huanglian Jiedu decoction (HLJDD) and its three major components alleviated the mRNA expression of the key mediators and inflammatory cytokines. |
|
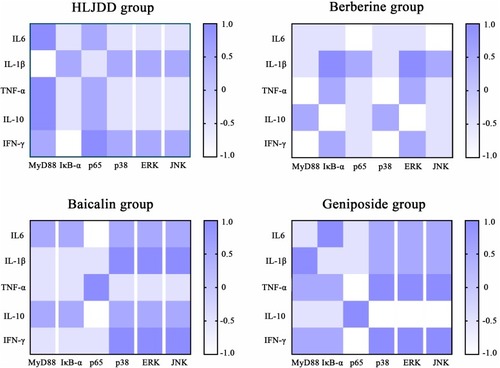
Correlation analysis of inflammatory cytokines and mediators of the TLR4/MyD88 signaling pathway in each drug treatment group. The color scale illustrates the magnitude of correlation between the examined indexes on the plot. |
|
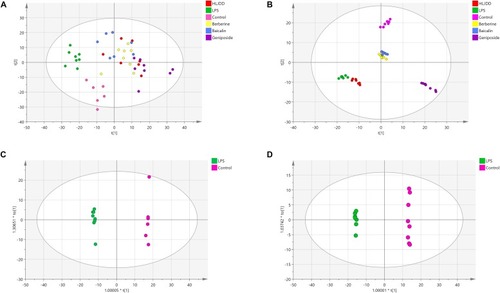
Principal component analysis (PCA) scores of zebrafish samples in six groups and orthogonal projection to latent structures discriminant analysis (OPLS-DA) scores in the control group and lipopolysaccharide (LPS) group. |
|
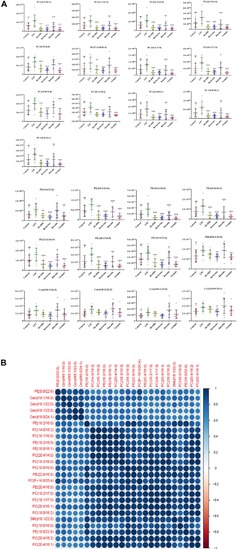
Multivariate statistical analysis of the lipopolysaccharide (LPS) group and control group. |
|
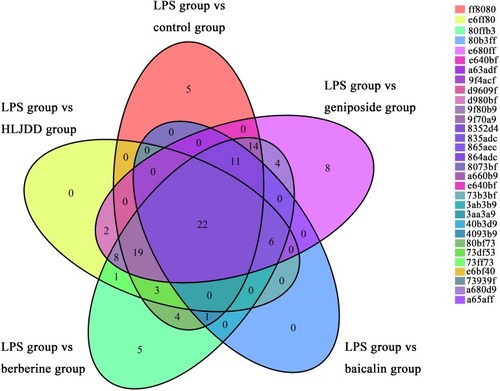
Venn diagram, in which each ellipse represents the potential lipid markers based on comparisons of the lipopolysaccharide (LPS) group |
|
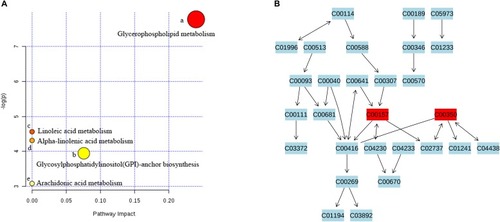
Metabolic pathway analysis and visualization of Huanglian Jiedu decoction (HLJDD) treatment. |
|
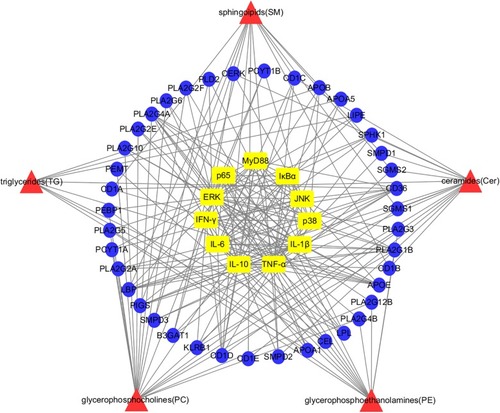
Network visualization analysis of “potential lipid markers-lipoproteins-TLR4/MyD88 signaling pathways” for anti-inflammatory effects of HLJDD. Red area: differentially expressed species of lipid markers; Blue area: corresponding lipoproteins of lipid markers; Yellow area: cytokines and proteins of the TLR4/MyD88 signaling pathways. |