- Title
-
Nitrogen deprivation induces triacylglycerol accumulation, drug tolerance and hypervirulence in mycobacteria
- Authors
- Santucci, P., Johansen, M.D., Point, V., Poncin, I., Viljoen, A., Cavalier, J.F., Kremer, L., Canaan, S.
- Source
- Full text @ Sci. Rep.
|
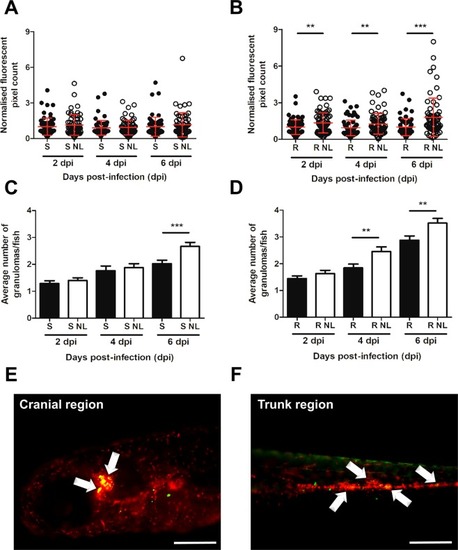
Nitrogen limitation induces hypervirulence in PHENOTYPE:
|
|
Nitrogen limitation results in increased bacterial burden and granuloma abundance. Transgenic zebrafish embryos harbouring red fluorescent macrophages ( PHENOTYPE:
|