- Title
-
Use of PTC124 for nonsense suppression therapy targeting BMP4 nonsense variants in vitro and the bmp4st72 allele in zebrafish
- Authors
- Krall, M., Htun, S., Slavotinek, A.
- Source
- Full text @ PLoS One
|
Results of In-cell ELISA assay, demonstrating BMP4 protein levels for the p.(Arg198*) and p.(Glu213*) variants after incubation of 293T/17 cells for 48 hours with 20 μM PTC124, compared to untreated cells transfected with the same construct. An increase in BMP4 protein level was seen with p.(Arg198*), but this was not significant (p value = 0.09). There was minimal increase in the construct containing p.(Glu213*) after treatment with 10 μM PTC124 and 20 μM PTC124. |
|
Representative examples of ventroposterior defects scored in PHENOTYPE:
|
|
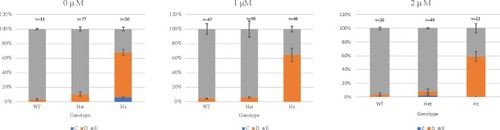
Larvae were not dechorionated and PTC124 treatment was started at 0 hours post fertilization (hpf). Analysis was performed at 72 hpf and the results are shown as the mean of 3 independent experiments, except for treatment with 2 μM PTC124, which is shown as the mean of 2 independent experiments. Larvae were scored as wildtype (E), mild reduction/deformity in ventral tail fin (D+ or D) or severe reduction/deformity in ventral tail fin (C). WT = wildtype, het = heterozygote, HZ = homozygote. Error bars show standard error of the mean. We tested for trend in the probability of tail fin defects across the increasing doses of 0 μM, 1.0 μM and 2.0 μM PTC124 treated, groups of homozygous larvae [ PHENOTYPE:
|
|
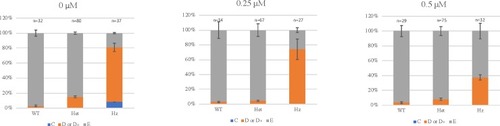
Larvae were dechorionated at 6–8 hours post fertilization (hpf) prior to starting PTC124 treatment. Analysis was performed at 72 hpf and the results are shown as the mean of 3 independent experiments. Larvae were scored as wildtype (E), mild reduction/deformity in ventral tail fin (D+ or D) or severe reduction/deformity in ventral tail fin (C). WT = wildtype, het = heterozygote, HZ = homozygote. Error bars show standard error of the mean. We tested for trend in the probability of tail fin defects across the increasing doses of 0 μM, 0.25 μM and 0.5 μM PTC124 treated, groups of homozygous larvae [ |
|
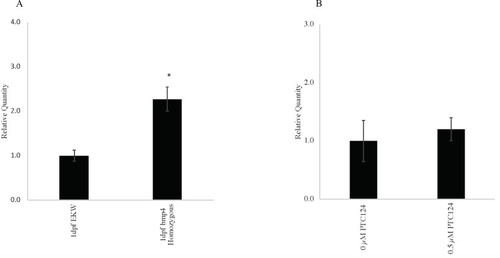
Fig 5A. Quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR) showing EXPRESSION / LABELING:
PHENOTYPE:
|