- Title
-
An enhancer polymorphism at the cardiomyocyte intercalated disc protein NOS1AP locus is a major regulator of the QT interval
- Authors
- Kapoor, A., Sekar, R.B., Hansen, N.F., Fox-Talbot, K., Morley, M., Pihur, V., Chatterjee, S., Brandimarto, J., Moravec, C.S., Pulit, S.L., Pfeufer, A., Mullikin, J., Ross, M., Green, E.D., Bentley, D., Newton-Cheh, C., Boerwinkle, E., Tomaselli, G.F., Cappola, T.P., Arking, D.E., Halushka, M.K., Chakravarti, A.
- Source
- Full text @ Am. J. Hum. Genet.
|
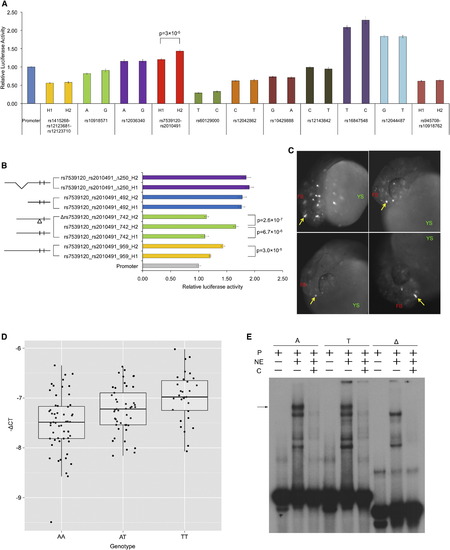
rs7539120 Is a Functional Variant Underlying QT Interval Association at the NOS1AP Locus (A) Firefly luciferase reporter enhancer/silencer assays in HL1 cells using alternate alleles/haplotypes for selected QT-interval-associated NOS1AP variants. Firefly luciferase expression is plotted relative to Renilla luciferase expression, normalized to the expression from empty vector (Promoter). Mean luciferase expression between alternate alleles/haplotypes constructs was compared with the t test and found to be significantly different between rs2010491 haplotypes, which also include rs7539120 alleles (p = 3 × 10−5). Error bars indicate SEM (n = 8). (B) Enhancer activity in rs7539120_rs2010491 haplotype is driven by rs7539120 and is dependent on the flanking sequence. Firefly luciferase reporter enhancer/silencer assays in HL1 cells using a deletion series derived from the 959 bp rs7539120_rs2010491 construct. In the construct design on left, small vertical lines represent the rs7539120 and rs2010491 SNPs, Δ represents the 11-base deletion encompassing rs7539120, 5′ deletions are represented by shorter lengths of the horizontal line, and an internal 250 bp deletion is represented by sloping lines. Error bars indicate SEM (n = 8). (C) rs7539120 acts as an in vivo enhancer. The rs7539120 risk haplotype construct injected (rs7539120_rs2010491_742_H2) at the 1–2 cell stage in developing zebrafish embryos drives transient reporter expression (enhanced-GFP; eGFP) in forebrain 24 hr postfertilization in ∼33% of injected embryos. Representative images from four different embryos with forebrain eGFP expression (white spots, strong expression indicated by yellow arrows) are shown. No eGFP expression was observed from the rs7539120 deletion (±5 bases) constructs (Δrs7539120_rs2010491_742_H2) (see Figure S4). Abbreviations are as follows: FB, forebrain; YS, yolk sac. (D) rs7539120 acts as a cardiac expression quantitative trait locus (eQTL). Box-whisker plots of NOS1AP mRNA expression in human left ventricle tissue from 131 heart failure subjects genotyped at rs7539120. The y axis shows –ΔCT used as a measure of expression. The risk allele T is associated with higher expression of NOS1AP (p = 4.72 × 10−5). (E) rs7539120 is bound by an uncharacterized protein(s) from HL1 cell nuclear extract. EMSAs using radiolabeled probes (P) containing nonrisk allele (A), risk allele (T), and deletion (Δ) of rs7539120 in presence or absence of HL1 cells nuclear extract (NE) and excess unlabeled competing probe (C). The black arrow indicates DNA-protein complex formed with both the A and T alleles containing probes, but lacking with the deletion probe. + indicates addition; - indicates absence. |
|
rs7539120 acts as an in vivo enhancer that overlaps one of the nos1apa endogenous expression domains. A) The rs7539120 non-risk haplotype construct, like the risk haplotype (see Figure 2), injected at 1-2 cell stage in developing zebrafish embryos drives transient reporter expression (enhanced GFP; eGFP) in forebrain 24 hours post fertilization in -25% of injected embryos. Representative images from three different embryos with forebrain eGFP expression (white spots, strong expression indicated by yellow arrows) are shown in top row. No eGFP expression was observed from the injections of rs 7539120 deletion ( +/- 5 bases) construct. Representative images from three different embryos with no eGFP expression are shown in bottom row. FB: forebrain; YS: yolk sac. B) (top) A brightfield image of a zebrafish embryo at 24 hours post fertilization. The red line marks the area used to dissect the head and the remainder of the body for expression analysis. (bottom) RT-PCR on cDNA prepared from either the entire embryos (Whole), or the head region (Head), or the remainder of the body (Remainder), as depicted in image above, using nos1apa and actb1 primers. nos1apa expression was observed in all three samples. actb 1 was used as house-keeping gene control. Control lane represents water samples used as negative control. M: DNA ladder; bp: base pair. EXPRESSION / LABELING:
|