- Title
-
The Isl1/Ldb1 Complex Orchestrates Genome-wide Chromatin Organization to Instruct Differentiation of Multipotent Cardiac Progenitors
- Authors
- Caputo, L., Witzel, H.R., Kolovos, P., Cheedipudi, S., Looso, M., Mylona, A., van IJcken, W.F., Laugwitz, K.L., Evans, S.M., Braun, T., Soler, E., Grosveld, F., Dobreva, G.
- Source
- Full text @ Cell Stem Cell
|
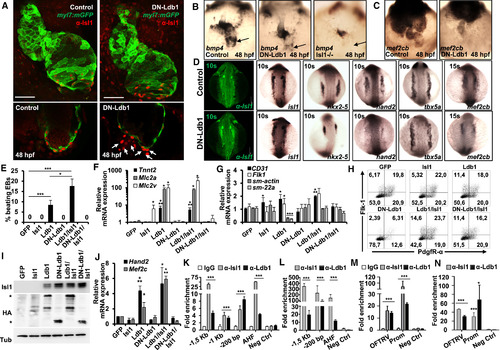
The Dimerization Domain of Ldb1 Is Required for Cardiomyocyte Differentiation (A) Confocal images of control and DN-Ldb1-overexpressing Tg(myl7:EGFP-HsHRAS)s883 zebrafish embryos at 48 hr post fertilization (hpf) stained with anti-GFP and anti-Isl1 (red) antibodies. Arrows indicate Isl1+ cells outside of the heart, which do not express the cardiomyocyte marker myl7. Scale bars, 50 μm. (B and C) In situ hybridization of control, DN-Ldb1-overexpressing, and isl1 mutant embryos at 48 hpf with bmp4 (B) and mef2cb (C) probes. The arrows indicate bmp4 expression at the sinus venosus. (D) Confocal images of control and DN-Ldb1-overexpressing embryos stained with anti-Isl1 antibody (leftmost panels) and in situ hybridization of control and DN-Ldb1-overexpressing embryos at 10–15 somite stages with isl1, nkx2.5, hand2, tbx5a, and mef2cb probes. (E) Percentage of beating d7 EBs derived from Ldb1−/− ESCs overexpressing either GFP alone (control) or GFP together with Isl1, Ldb1, DN-Ldb1, or these in different combinations. (F and G) Relative mRNA expression analysis of cardiomyocyte (F), smooth muscle, and endothelial marker (G) genes in d7 EBs. (H) FACS analysis of Flk-1 and PdgfR-α expression in d3.75 Ldb1−/− EBs overexpressing either GFP alone or GFP together with Isl1, Ldb1, DN-Ldb1, or these in different combinations. (I) Western blot analysis of total protein extracts of d5 EBs. (J) Relative mRNA expression of Mef2c and Hand2 in d4 EBs. (K and L) ChIP of nuclear extracts from d5 EBs (K) and pools of dissected SHF from E8–9 embryos (L) using anti-Isl1 and anti-Ldb1 antibodies or IgG as control. PCRs were performed using primers flanking conserved Isl1 binding sites in the Mef2c promoter and the AHF enhancer. Fold enrichment values for EBs were calculated relative to IgG control and for embryos relative to a genomic region that does not contain conserved Isl1 binding sites. Data are mean ± SEMs, n = 3. (M and N) ChIP of nuclear extracts from d5 EBs (M) and pools of dissected SHF from E8–9 embryos (N) using anti-Isl1 and anti-Ldb1 antibodies or IgG as a control. Data are mean ± SEMs, n = 3. Data in (E)–(G) and (J) are mean ± SEMs, n = 3. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.005. See also Figures S3–S7. |
|
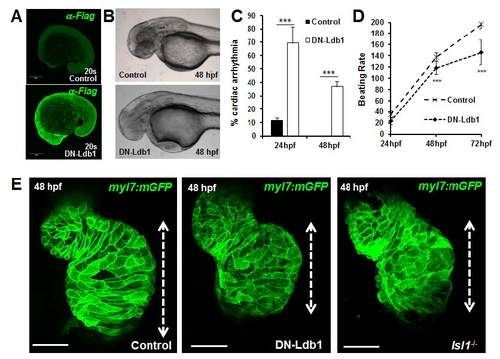
Cardiac morphogenesis defects in zebrafish embryos overexpressing DN-Ldb1. (A) Confocal images of control and FLAG-HA-DN-Ldb1 mRNA injected zebrafish embryos, stained with anti-FLAG antibody at 20 somites. (B) Control or DNLdb1 mRNA injected Tg(myl7:EGFP-HsHRAS)s883 embryos at 48 hpf. Lateral view, anterior to the left. (C-D) Percentage of embryos with cardiac arrhythmia (C) and analysis of the number of heart beats per minute (D) measured at 24, 48 and 72 hpf in control and DN-Ldb1 overexpressing zebrafish embryos. (E) Confocal images of control, DN-Ldb1 overexpressing and Isl1 mutant Tg(myl7:EGFP-HsHRAS)s883 hearts, showing shortening of the atrium (dotted lines) in DN-Ldb1 overexpressing and isl1 mutant embryos. |