- Title
-
Zebrafish small molecule screens: Taking the phenotypic plunge
- Authors
- Williams, C.H., Hong, C.C.
- Source
- Full text @ Comput Struct Biotechnol J
|
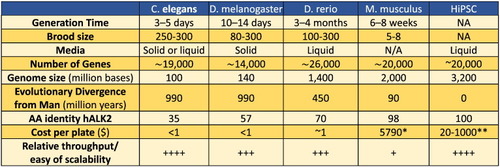
Comparison of model organisms used in phenotypic screens. Commonly accepted numbers for generation time and brood size are listed, along with media for animal maintenance, evolutionary divergence, gene number and genome size |
|
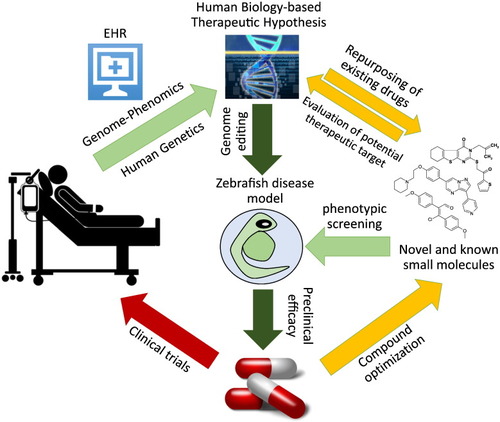
Proposed zebrafish phenotypic screens incorporating human genome–phenome information to accelerate therapeutic discovery. Human genome–phenome information provided by electronic health record (EHR)-coupled DNA database and by human genetic diseases studies drive formulation of therapeutic hypotheses (“human biology-based therapeutic hypotheses”). To test these hypotheses, zebrafish models of human genetic diseases are generated by genomic editing and employed in phenotypic screen for novel or known compounds which ameliorate the disease phenotypes. These compounds are then advanced for further development, including compound optimization and testing in appropriate preclinical disease models. Alternatively, a target-agnostic morphology-based screen is carried out. Subsequently, targets of hit compounds identified, and each target evaluated |