- Title
-
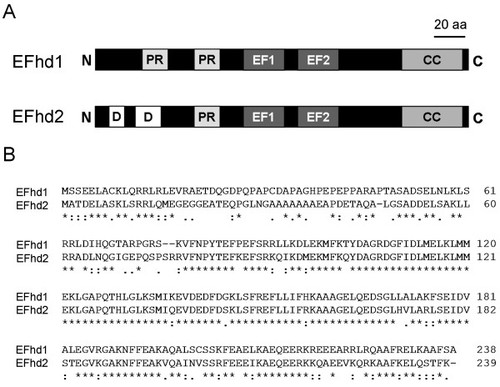
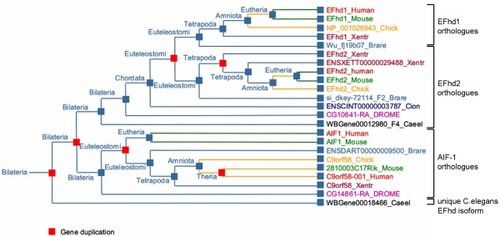
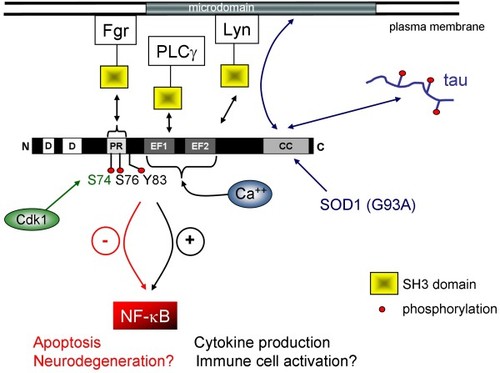
Fraternal twins: Swiprosin-1/EFhd2 and Swiprosin-2/EFhd1, two homologous EF-hand containing calcium binding adaptor proteins with distinct functions
- Authors
- Dütting, S., Brachs, S., and Mielenz, D.
- Source
- Full text @ Cell Commun. Signal.
|
|
|
|
|
|
|
|