- Title
-
Analyses of zebrafish and Xenopus oocyte maturation reveal conserved and diverged features of translational regulation of maternal cyclin B1 mRNA
- Authors
- Zhang, Y., and Sheets, M.D.
- Source
- Full text @ BMC Dev. Biol.
|
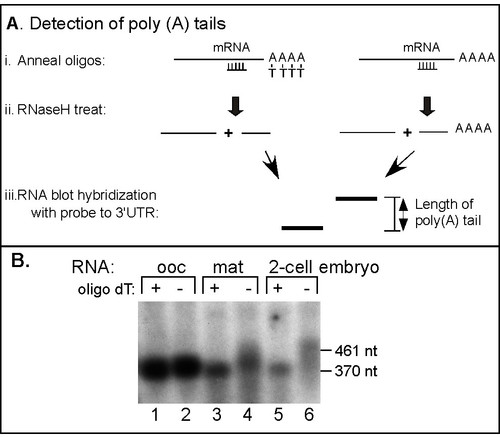
The endogenous zebrafish cyclinB1 mRNA is polyadenylated during oocyte maturation and early embryogenesis. (A) Diagram of oligonucleotide/RNaseH treatment for analysis of mRNA poly (A) tails. i.) RNA samples were hybridized to a DNA oligonucleotide complementary to a region of the open reading frame close to the 3′UTR. Half of each sample was also hybridized to oligo/dT. ii.) Treatment with RNaseH cleaved the RNA in the region of the RNA/DNA hybrid. In the sample with oligo/dT RNaseH cleaved the poly (A) tail. iii.) Treated RNA samples were analyzed by high resolution RNA blot hybridization and the sizes of the 3′UTR fragments with and without poly (A) indicates the length of poly (A) tail present. (B) Analysis of the endogenous zebrafish cyclinB1 mRNA 3′UTR following oligonucleotide/RNaseH treatment. Total RNAs from zebrafish oocytes, mature oocytes and two-cell embryos were hybridized with a DNA oligonucleotide complimentary to the zebrafish cyclin B1 mRNA's 3′UTR nucleotides -393 to -372 relative to poly(A) addition site at +1. Half of each sample was also hybridized with oligo dT (lanes 1, 3, 5). All samples were then treated with RNaseH and analyzed by RNA blot hybridization using a radio labeled probe corresponding to the 3′UTR of zebrafish CyclinB1 mRNA (-199 to +1). The positions of RNA markers are indicated. |
|
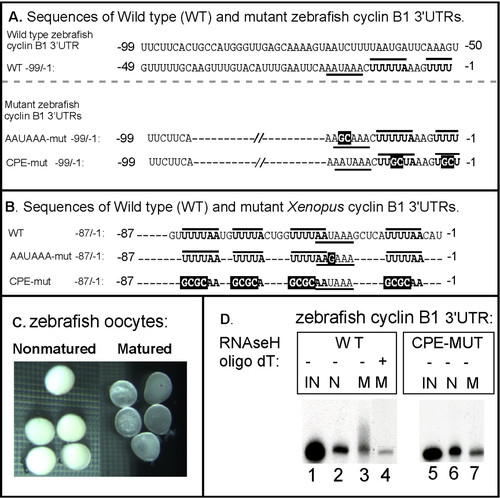
Polyadenylation of the zebrafish cyclin B1 mRNA 3′UTR depended upon U-rich CPE sequences. (A) Sequences of wild-type, AAUAAA mutant and U-rich mutant zebrafish cyclin B1 3′UTRs. The zebrafish cyclin B1 3′UTR is 199 nucleotides in length and the last 99 nucleotides are shown. U-rich putative CPE elements adjacent to AAUAAA are overlined and bolded while the AAUAAA sequence is underlined. Mutated sequences are white on black. Other U-rich sequences are present in the zebrafish cyclin B1 3′UTR, but their distance from AAUAAA makes it unlikely that they function as CPEs. (B) Sequences of wild-type, AAUAAA mutant and CPE mutant Xenopus cyclinB1 3′UTRs. Mutated sequences are white on black, CPE elements are overlined and bolded and AAUAAA elements are underlined. The numbers indicate positions of each nucleotide relative to the poly (A) addition site. (C) Zebrafish oocytes and matured oocytes. (D) Zebrafish cyclin B1 3′UTR is sufficient to direct polyadenylation during oocyte maturation, and the CPEs are required. P32-labeled RNA consisting of the zebrafish cyclin B1 mRNA 3′UTR was injected into 50–100 zebrafish oocytes and some oocytes were matured. RNA from injected non-mature oocytes (N) or injected matured oocytes (M) were analyzed by 4% denaturing PAGE and autoradiography. IN: Uninjected RNA. N: RNA from non-mature oocytes. M: RNA from mature oocytes. As controls, half of the total RNAs extracted from WT 3′UTR RNA injected matured oocytes were subjected to oligo dT/RNaseH treatment prior to denaturing PAGE (lane 4). The size reduction after oligo dT/RNaseH treatment indicated that the 3′UTR RNA was polyadenylated. |
|
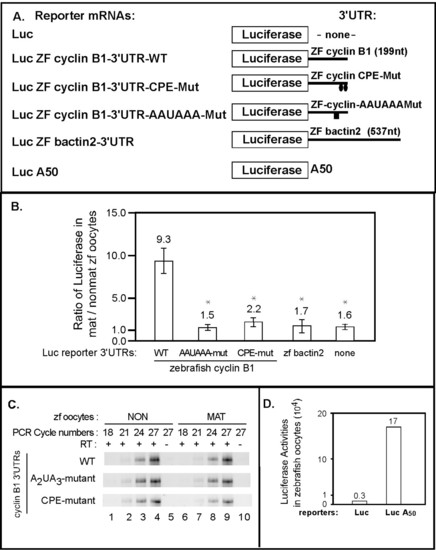
The 3′UTR of zebrafish cyclinB1 mRNA is sufficient to activate translation during zebrafish oocyte maturation and this activation requires the CPE and AAUAAA sequences. (A) Schematic of various luciferase reporter mRNAs with different 3′UTRs (ZF-zebrafish). (B) The zebrafish cyclin B1 3′UTR directs translational activation during oocyte maturation and activation requires the putative CPE elements and AAUAAA element. Luciferase reporter mRNAs with various zebrafish cyclinB1 3′UTRs were injected into oocytes. Some of the injected oocytes were matured. Luciferase activity was measured in extracts prepared from injected oocytes and injected matured oocytes. The ratio of luciferase in matured versus non-matured oocytes was plotted for comparison. The mat/non-mature ratios shown are represented as mean +/- SEM of at least three independent experiments. Statistical analysis (P values) was performed by one-way ANOVA. * P < 0.05. Each reporter containing zebrafish cyclin B1 mutant 3′UTR, bactin2 3′UTR or no 3′UTR was compared to the reporter with zebrafish cyclin B1 WT 3′UTR. (C) Luciferase reporters were equally stable in mature vs. non-mature oocytes. Total RNA was extracted from mature and non-matured oocytes injected with each zebrafish cyclin B1 3′UTR reporter mRNA. Equal amounts of total RNA from each sample were utilized in quantitative RT-PCR using luciferase-specific primers. Product formation was monitored with increasing cycles of amplification and analyzed by agarose gel electrophoresis. (D) The presence of a poly (A) tail is sufficient to stimulate mRNA translation in zebrafish oocytes. Luciferase reporter mRNAs (Luc vs. LucA50) were injected into 20–30 fully-grown zebrafish oocytes and assayed for luciferase activity after 6 hours. This experiment was repeated twice and shown here are the absolute values of luciferase activity from one representative experiment. |
|
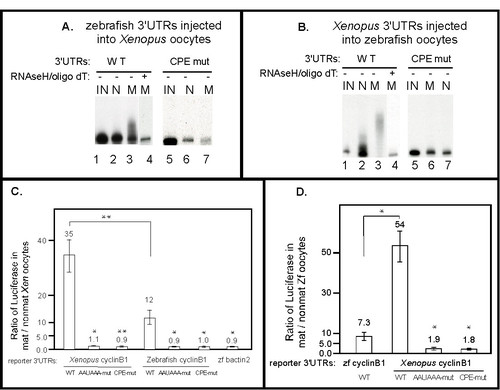
Directing polyadenylation and activating translation during oocyte maturation are evolutionarily conserved functions of cyclin B1 3′UTRs. (A) The zebrafish cyclinB1 3′UTR is sufficient to direct polyadenylation during Xenopus oocyte maturation, and the CPEs are required. Each P32-labeled 3′UTR RNA was injected into 30–50 Xenopus oocytes and some oocytes were matured. RNA isolated from injected non-mature oocytes or injected matured oocytes were analyzed by 4% denaturing PAGE. IN: Uninjected RNA. N: RNA from non-mature oocytes. M: RNA from mature oocytes. Half of the RNA from WT 3′UTR injected mature oocytes was treated with oligo dT/RNaseH prior to analysis (lane 4). The size reduction after oligo dT/RNaseH treatment (lane 4 versus lane 3) indicated that the WT 3′UTR RNA was polyadenylated. (B) The Xenopus cyclinB1 3′UTR is sufficient to direct polyadenylation during zebrafish oocyte maturation, and the CPEs are required. Each P32-labeled 3′UTR RNA was injected into 50–100 zebrafish oocytes and some oocytes were matured. RNA from injected non-matured oocytes or injected matured oocytes were analyzed by 4% denaturing PAGE. IN: Uninjected RNA. N: RNA from non-mature oocytes. M: RNA from mature oocytes. Half of the RNA extracted from WT 3′UTR injected matured oocytes was treated with oligo dT/RNaseH prior to analysis (lane 4). The size reduction after oligo dT/RNaseH treatment indicated that the WT 3′UTR RNA was polyadenylated (compare lanes 3 and 4). (C) The zebrafish cyclinB1 3′UTR is sufficient to activate translation during Xenopus oocyte maturation and the AAUAAA and CPE sequences are required for this activation. Luciferase reporter RNAs with various 3′UTRs were injected into 30–40 Xenopus oocytes and half were matured with progesterone. The ratios of luciferase activities from mature vs. non-mature oocytes were calculated and graphed as mean +/- SEM from at least three independent experiments. Statistical analysis was performed by one-way ANOVA. * P < 0.05, ** P < 0.01. Each Xenopus mutant 3′UTR was compared to Xenopus WT 3′UTR, each zebrafish mutant cyclin B1 3′UTR or WT bactin2 3′UTR was compared to zebrafish WT cyclin B1 3′UTR. The mature/non-mature ratio of Xenopus cyclin B1 3′UTR is significantly different from that of zebrafish cyclin B1 3′UTR (D) The Xenopus Cyclin B1 3′UTR is sufficient to activate translation during zebrafish oocyte maturation and the AAUAAA and CPE sequences are required for this activation. Luciferase reporter RNAs with various 3′UTRs were injected into 50–100 zebrafish oocytes and half were matured with hormone. The ratio of luciferase activities from mature versus non-mature oocytes were calculated and graphed as the mean values +/- SEM from at least three independent experiments. Statistical analysis was performed by one-way ANOVA. * P < 0.05. Each mutant Xenopus reporter was compared to WT Xenopus reporter. The mature/non-mature ratio of zebrafish cyclin B1 3′UTR is significantly different from that of Xenopus cyclin B1 3′UTR. |