- Title
-
Morphogenesis is transcriptionally coupled to neurogenesis during peripheral olfactory organ development
- Authors
- Aguillon, R., Madelaine, R., Aguirrebengoa, M., Guturu, H., Link, S., Dufourcq, P., Lecaudey, V., Bejerano, G., Blader, P., Batut, J.
- Source
- Full text @ Development
|
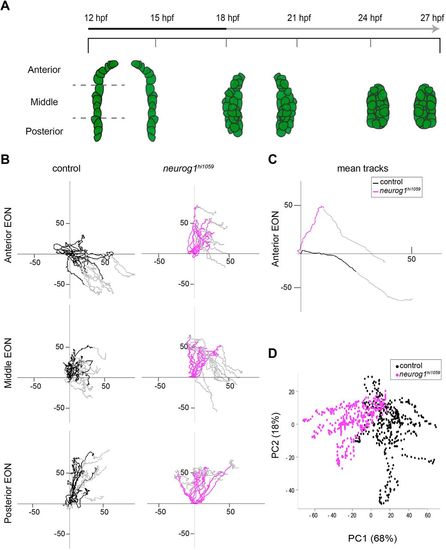
Oriented cell movements are affected in neurog1hi1059 mutant embryos during olfactory cup formation. (A) Graphic representation of the morphogenesis of olfactory cup from 12?hpf to 27?hpf, showing a dorsal view of the three olfactory stages: olfactory territory (12?hpf), olfactory placode (18?hpf) and olfactory cup (24?hpf). EON progenitors are represented in green as visualised with the Tg(-8.4neurog1:gfp) transgene. At 12?hpf, the ?8.4neurog1:GFP+ placodal domain can be divided into anterior, middle and posterior regions. The early (12-18?hpf; black) and late (18-27?hpf; grey) phases of morphogenesis are noted in the time line. (B) Tracks showing migration of EON of control (black) or neurog1hi1059 mutant (magenta) embryos. Twelve tracks are represented (two cells each from the left and right of three embryos) for each of the anterior, middle and posterior domains of the developing cup indicated in A. The origin of the tracks has been arbitrarily set to the intersection of the x/y-axes and the early (coloured) and late (grey) phases of migration have been highlighted. (C) Mean tracks for anterior EON of control (black) or neurog1hi1059 mutant (magenta) embryos. (D) Pairwise PCA scatterplots of morphogenetic parameters extracted from the datasets corresponding to the tracks in C. The major difference between control and neurog1hi1059 mutant embryos (PC1) corresponds to the antero-posterior axis. |
|
Morphogenetic defects in cxcr4bt26035 and cxcl12at30516 mutant embryos resemble those of neurog1hi1059. (A) Tracks showing migration of anterior EON from control (black), cxcr4bt26035 (blue) and cxcl12at30516 (green) embryos. Twelve anterior tracks are represented (two cells each from the left and right of three embryos). The origin of the tracks has been arbitrarily set to the intersection of the x/y-axes and the early (coloured) and late (grey) phases of migration have been highlighted. (B) Mean tracks showing migration of anterior EON of control (black), neurog1hi1059 (magenta), cxcr4bt26035 (blue) and cxcl12at30516 (green) mutant embryos. (C) Pairwise PCA scatterplots of morphogenetic parameters extracted from the datasets corresponding to the tracks in B. The major difference between control, neurog1hi1059, cxcr4bt26035 and cxcl12at30516 mutant embryos (PC1) corresponds to the antero-posterior axis. (D) Clustering analysis of morphogenetic parameters extracted from the datasets presented in B and analysed in C. One cluster, k1, contains almost exclusively control cells (black), whereas cells from neurog1hi1059 (magenta), cxcr4bt26035 (blue) and cxcl12at30516 (green) mutant embryos clustered together in k2, k3 and k4. |
|
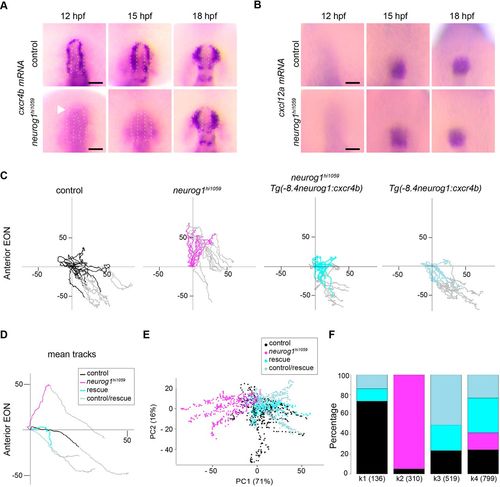
Fig. 3. Cxcr4b is the predominant downstream effector of Neurog1 during olfactory cup morphogenesis. (A) cxcr4b whole-mount in situ hybridisation at 12, 15 and 18?hpf in control and neurog1hi1059 mutant embryos. cxcr4b expression is dramatically reduced or absent in EON progenitors at 12 and 15?hpf in neurog1hi1059 mutant embryos (white dotted lines) but from 18?hpf the expression of cxcr4b recovers. (B) cxcl12a whole-mount in situ hybridisation at 12, 15 and 18?hpf in control and neurog1hi1059 mutant embryos, in which cxcl12a expression is not affected. (C) Tracks showing migration of anterior EON of control (black) embryos, neurog1hi1059 mutant embryos (magenta), neurog1hi1059 mutant embryos carrying the Tg(-8.4neurog1:cxcr4b) transgene (cyan) and control embryos carrying Tg(-8.4neurog1:cxcr4b) (light blue). Twelve anterior tracks are represented (from four embryos). The origin of the tracks has been arbitrarily set to the intersection of the x/y-axes and the early (coloured) and late (grey) phases of migration have been highlighted. (D) Mean tracks showing migration of anterior EON of the tracks in C. (E) Pairwise PCA scatterplots of morphogenetic parameters extracted from the datasets presented in D. The major difference between control, neurog1hi1059, and control or neurog1hi1059 with the rescue transgene (PC1) corresponds to the antero-posterior axis. (F) Clustering analysis of morphogenetic parameters extracted from the datasets presented in D and analysed in E. One cluster, k2, contains virtually exclusively neurog1hi1059 cells (magenta), whereas cells from control (black), rescue (cyan) and control/rescue (light blue) embryos clustered together in k1, k3 and k4. Scale bars: 100 µm. |
|
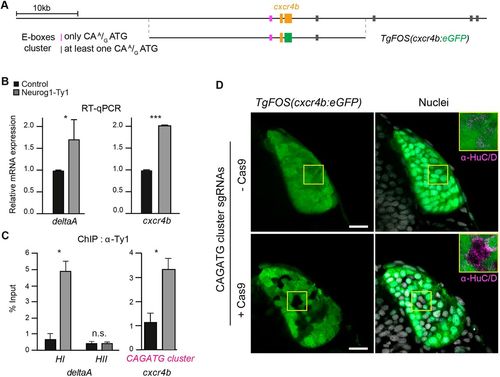
Neurog1 directly controls cxcr4b expression via an upstream cis-regulatory module (CRM). (A) Schematic of the cxcr4b locus, indicating the position of exons of the cxcr4b gene (orange) and E-box clusters, which are colour-coded depending on the nature of the E-box sequences. Also presented is the position of the genomic sequences found in the TgFOS(cxcr4b:eGFP) transgene. (B) qPCR analysis of the effect of Neurog1-Ty1mRNA mis-expression on the relative mRNA levels of the known Neurog1 target gene deltaA and cxcr4b. A significant increase in expression is detected for both genes. (C) ChIP using an antibody against Ty1 and chromatin prepared from 15?hpf embryos mis-expressing Neurog1-Ty1 mRNA (grey). Control (black) represents ChIP with IgG alone. (D) Single confocal sections of TgFOS(cxcr4b:eGFP) embryos at 24?hpf, showing eGFP expression in the olfactory cups, and either HuC/D expression or nuclear labelling (TOPRO). Embryos were injected with an sgRNA pair flanking the E-box-containing CRM at the cxcr4b locus plus or minus Cas9 as a control. Insets show HuC/D expression in both conditions. Data are meanħs.e.m. *P=0.01, ***P=0.0001 (two-tailed Student's t-test). n.s., not significant. Scale bars: 20 µm. |