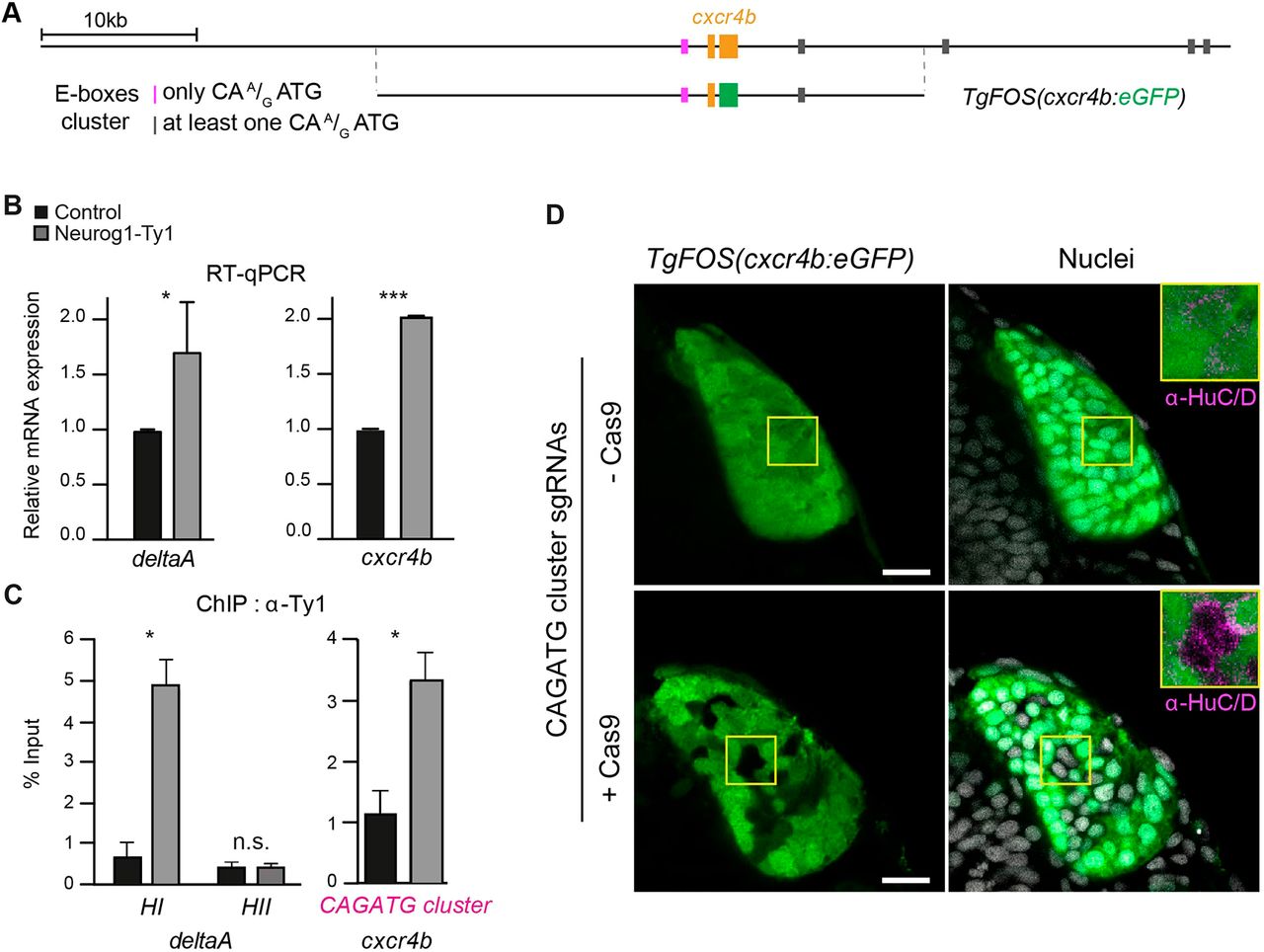
Fig. 4 Neurog1 directly controls cxcr4b expression via an upstream cis-regulatory module (CRM). (A) Schematic of the cxcr4b locus, indicating the position of exons of the cxcr4b gene (orange) and E-box clusters, which are colour-coded depending on the nature of the E-box sequences. Also presented is the position of the genomic sequences found in the TgFOS(cxcr4b:eGFP) transgene. (B) qPCR analysis of the effect of Neurog1-Ty1mRNA mis-expression on the relative mRNA levels of the known Neurog1 target gene deltaA and cxcr4b. A significant increase in expression is detected for both genes. (C) ChIP using an antibody against Ty1 and chromatin prepared from 15 hpf embryos mis-expressing Neurog1-Ty1 mRNA (grey). Control (black) represents ChIP with IgG alone. (D) Single confocal sections of TgFOS(cxcr4b:eGFP) embryos at 24 hpf, showing eGFP expression in the olfactory cups, and either HuC/D expression or nuclear labelling (TOPRO). Embryos were injected with an sgRNA pair flanking the E-box-containing CRM at the cxcr4b locus plus or minus Cas9 as a control. Insets show HuC/D expression in both conditions. Data are mean±s.e.m. *P=0.01, ***P=0.0001 (two-tailed Student's t-test). n.s., not significant. Scale bars: 20 µm.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Development