- Title
-
Gene evolution and gene expression after whole genome duplication in fish: the PhyloFish database
- Authors
- Pasquier, J., Cabau, C., Nguyen, T., Jouanno, E., Severac, D., Braasch, I., Journot, L., Pontarotti, P., Klopp, C., Postlethwait, J.H., Guiguen, Y., Bobe, J.
- Source
- Full text @ BMC Genomics
|
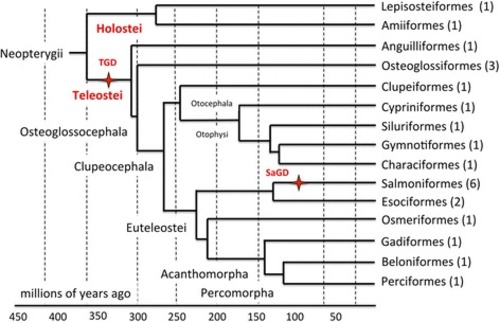
Phylogenetic tree of the PhyloFish species. Cladogram showing phylogenetic relationships among ray-finned fish analyzed in the present study. Tree topology was adapted from [ |
|
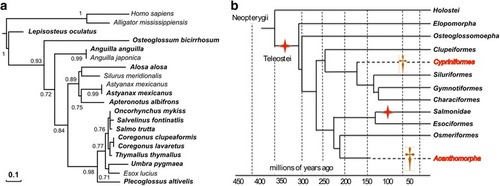
Stra8 proposed gene evolution in teleosts following the TGD WGD. Maximum-likelihood phylogeny of Stra8 ( |
|
Tissue expression profiles of |
|
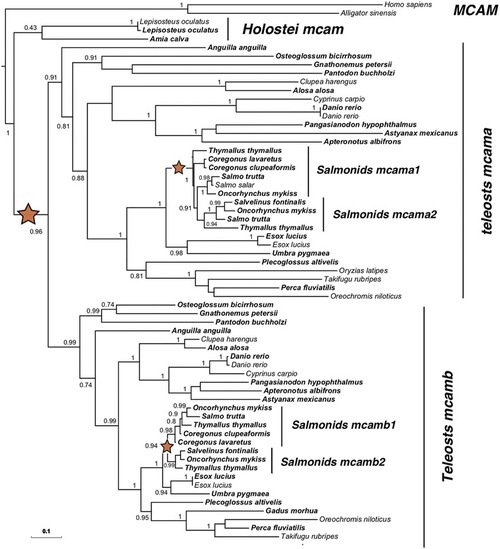
Phylogeny of Mcam in teleosts following the TGD and SaGD WGDs. Maximum-likelihood phylogeny of Mcam was performed using the PhyML software [ |