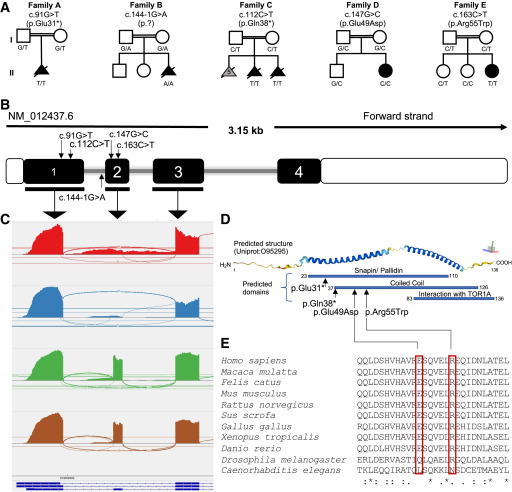
Fig. 1 Bi-allelic variants in SNAPIN are associated with a prenatal neuroanatomical syndrome in five unrelated pedigrees (A) Five pedigrees (families A–E) harbor bi-allelic SNAPIN variants. Homozygous variants are named according to canonical transcript GenBank: NM_012437.6. Unfilled symbols, healthy individuals; filled black symbols, affected individuals; gray fill, similar phenotype in previous pregnancy but DNA sample was not available; double line, consanguinity; squares, male; circles, females; triangles, prenatal affected individuals. (B) Schematic of the SNAPIN canonical transcript (GenBank: NM_012437.6). Filled and unfilled regions represent exons (with number in white font) and untranslated regions, respectively; gray line, introns. Approximate position of the identified variants is marked with small arrows. The figure is not to scale. (C) Sashimi plot showing RNA sequencing data generated from primary fibroblasts from family B harboring the c.144−1G>A change at the exon 2 splice acceptor site. Red, proband with nonsense-mediated decay (NMD) blocking; blue, proband without NMD blocking; green and brown, unrelated controls. (D) Schematic of predicted protein structure and domains of SNAPIN. Top, predicted helical structure generated with AlphaFold.23,24 Colors represent model confidence per residue ranging from 1 to 100; dark blue, very high confidence score (>90); sky blue, high confidence (70–90); yellow, low confidence (50–70); and orange, very low confidence (<50). Axis on top right, approximate plane of view where red, green, and blue bars denote the x, y, and z axes, respectively. Bottom: predicted SNAPIN domains (DRSC/TRiP Functional Genomics Resources)25 and location of variants. (E) Multiple sequence alignment of missense variants across species. “:” indicates similar amino acids, and asterisk (∗) denotes conserved sequence identity.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Am. J. Hum. Genet.