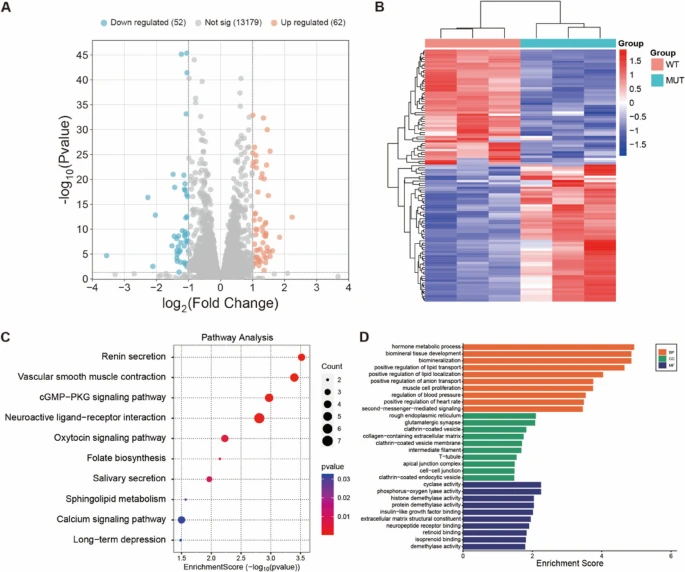
Fig. 6 A Volcano plot of differentially expressed genes (n = 3 technical replicates per group, single RNA-seq experiment). Green dots represent down-regulated genes, and red dots represent up-regulated genes (|log2FC|≥ 1 and adjusted p ≤ 0.05). B Heatmap showing the expression levels of transcripts in samples from the DPSCs-MUT and DPSCs-WT (n = 3 technical replicates per group). Red color refers to upregulation, and blue color refers to downregulation of gene transcription. C KEGG pathway analysis ranked the top 10 KEGG pathways (|log2FC|≥ 1 and adjusted p ≤ 0.05, based on 3 technical replicates per group). D Go results of biological process, cellular component, and molecular function (enrichment analysis based on differentially expressed genes with adjusted p ≤ 0.05, using 3 technical replicates per group). RNA sequencing was performed using Illumina platform with > 20 million clean reads per sample. Differential expression analysis was conducted using DESeq2 with Benjamini-Hochberg correction for multiple testing. Statistical significance: *adjusted p < 0.05, **adjusted p < 0.01, ***adjusted p < 0.001
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ BMC Oral Health