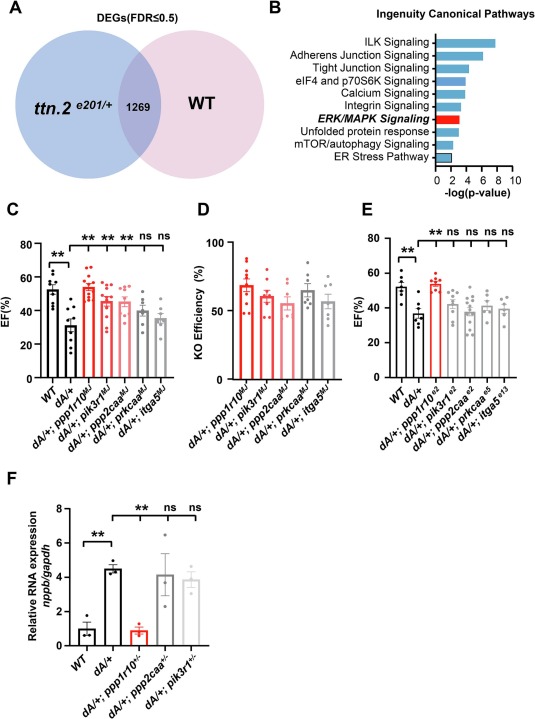
Fig. 3 An F0-based genetic screen of DEGs in Erk signaling pathway identified ppp1r10 as another therapeutic modifier. (A) Transcriptome analysis of ventricles from ttnae201/+ and WT at 6 months identified 1269 DE genes. (B) IPA analysis identified candidate pathways that warrant further investigation. (C) Quantification of ejection fraction in F0 adult zebrafish at 3 months via high-frequency echocardiography. (n ≥ 6 per group). (D) Quantification of representative knock out scores in F0 fish at 3 months. (E) Quantification of ejection fraction in F1 zebrafish at 3 months via high-frequency echocardiography. (n ≥ 6 per group). (F) Evaluation of nppb gene transcripts expression by quantitative RT-PCR in hearts from 6 months adult zebrafish. n ≥ 3. One-way ANOVA was used to compare multiple groups for each mutation. Data are presented as mean ± SEM, **p < 0.01. WT, wild type, ns, non-significant.
Reprinted from Journal of Molecular and Cellular Cardiology, , Yan, F., Wang, W., Moossavi, M., Zhu, P., Odell, N., Xu, X., Deregulated nutrient response in ttntv cardiomyopathy can be repaired via Erk inhibition for cardioprotective effects, , Copyright (2025) with permission from Elsevier. Full text @ J. Mol. Cell. Cardiol.