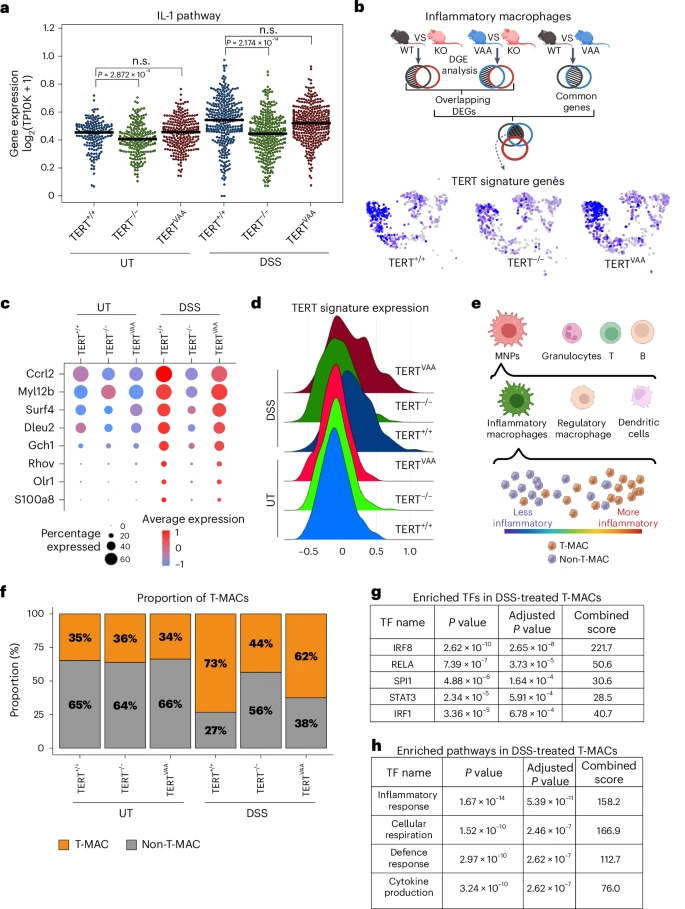
Fig. 5 Single-cell analysis revealed a TERT-driven myeloid subpopulation. a, The swarm plot shows the average expression of IL-1 pathway genes in inflammatory macrophage clusters of DSS-treated or untreated TERT+/+, TERTVAA and TERT−/− groups. (UT TERT+/+ n = 164, UT TERT−/− n = 217, UT TERTVAA n = 252, DSS TERT+/+ n = 314, DSS TERT−/− n = 341, DSS TERTVAA n = 305 cells). The line indicates the median expression. P values were calculated by two-sided Mann–Whitney test (n.s., not significant). b, Simplified schematic approach of TERT signature gene set identification. Multiple pair comparisons between untreated and DSS-treated TERT+/+ (WT), TERTVAA (VAA) and TERT−/− (KO) inflammatory macrophages revealed genes that are significantly upregulated in DSS-treated TERT+/+ and TERTVAA but not in TERT−/−. UMAPs show the average expression of TERT signature genes in MNPs of TERT+/+, TERTVAA and TERT−/− groups. c, The feature plot shows the TERT signature genes across genotypes with and without DSS treatment. d, The average expression of TERT signature genes in TERT+/+, TERTVAA and TERT−/− groups with or without DSS treatment. e, A schematic representation of T-MAC identification. Cells highly expressing TERT signature genes among inflammatory macrophages are annotated as T-MACs and lowly expressing ones as non-T-MAC. f, The bar plot shows the proportion of T-MAC and non-T-MAC cells in inflammatory macrophages of TERT+/+, TERTVAA and TERT−/− groups with or without DSS treatment. g, The list of transcription factors (TF) enriched in controlling DEGs that are downregulated in TERT−/− T-MAC cells than the TERT+/+ T-MACs upon DSS treatment. P values were calculated by two-sided Fisher exact test. h, The list of enriched pathways based on the upregulated genes in DSS-treated TERT+/+ T-MACs compared with untreated TERT+/+ T-MACs. P values were calculated by two-sided Fisher exact test. P values for a: UT-TERT+/+-versus-UT-TERT−/−, P = 2.872 × 10−4; DSS-TERT+/+-versus-DSS-TERT−/−, P = 2.174 × 10−14. Panels b and e created with BioRender.com. Source numerical data are available in Source data.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Nat. Cell Biol.