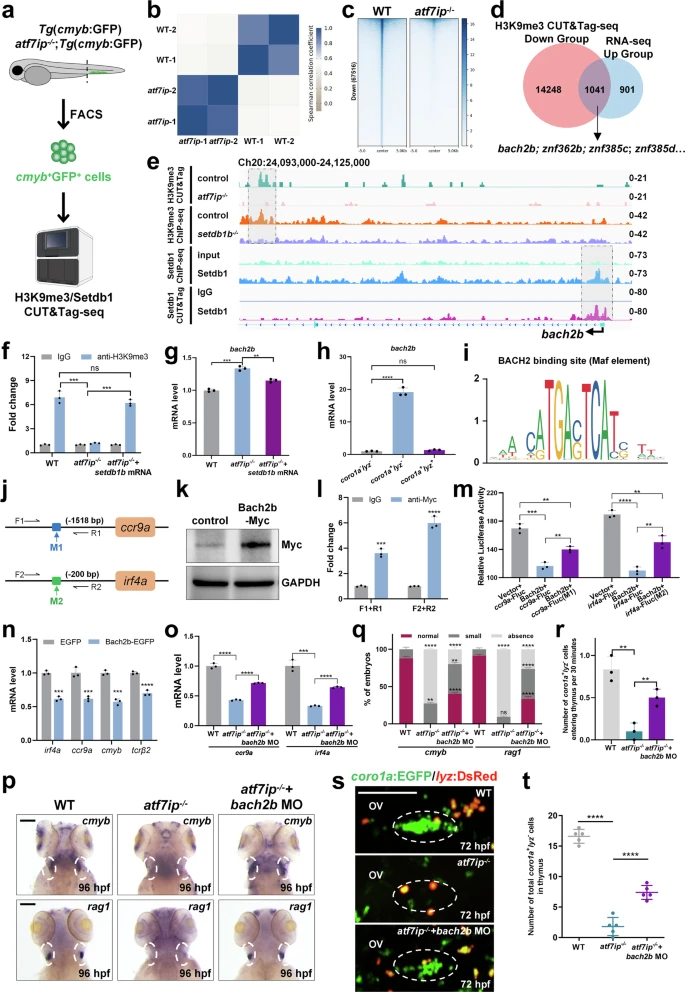
Fig. 6 Atf7ip cooperates with Setdb1 to target bach2b. a Schematic workflow of H3K9me3 and Setdb1 CUT&Tag-seq from FACS-sorted cmyb+ cells in WT and atf7ip−/− mutants at 72 hpf. b Heatmap of replicate correlations for H3K9me3 CUT&Tag in WT and atf7ip−/− mutants. c Heatmap for differentially enriched H3K9me3 peaks in whole genomes of WT and atf7ip−/− mutants. d Venn diagram displaying overlaps between gene-sets with reduced H3K9me3 and increased RNA-seq levels in atf7ip−/− mutants. e IGV tracks of H3K9me3 CUT&Tag-seq, H3K9me3 ChIP-seq, Setdb1 ChIP-seq and Setdb1 CUT&Tag-seq at bach2b locus in WT, atf7ip−/− and setdb1b−/− mutants. f CUT&Tag-qPCR of H3K9me3 levels at bach2b in WT, atf7ip−/−, setdb1b mRNA-injected atf7ip−/− mutants, n = 3. g qPCR of bach2b in WT, atf7ip−/− and setdb1b mRNA-injected atf7ip−/− mutant at 96 hpf, n = 3. h qPCR of bach2b in coro1a-lyz-, coro1a+lyz- and coro1a+lyz+ cells at 96 hpf, n = 3. i JASPAR database displaying BACH2 binding site (Maf element) in ccr9a and irf4a promoters. j CUT&Tag and luciferase reporter assay schematic with primers for ccr9a and irf4a promoters. k Immunoblot of Myc in bach2b-myc mRNA-injected embryos, n = 3. l qPCR of DNA fragments obtained from CUT&Tag assay with primers (F1, R1 or F2, R2), n = 3. m Luciferase activity in HEK293T cells after co-transfection of pGL3-ccr9a-Fluc, Bach2b, and pGL3-ccr9a-Mutant1 (M1), or pGL3-irf4a-Fluc, Bach2b, and pGL3-irf4a-Mutant2 (M2), n = 3. n qPCR of irf4a, ccr9a, cmyb, tcrβ2 in whole embryos of EGFP or bach2b-EGFP mRNA-injected WT (EGFP or Bach2b-EGFP) at 72 hpf, n = 3. o qPCR of ccr9a and irf4a in whole embryos of WT, atf7ip−/− and atf7ip−/− mutants injected with bach2b MO (8 ng per embryo) at 72 hpf, n = 3. p, q WISH displaying cmyb and rag1 expression in thymus (white circles) of WT, atf7ip−/− and bach2b MO-injected atf7ip−/− mutants at 96 hpf. Red, dark gray and light gray squares indicate normal (cmyb/rag1: 45-90), small (cmyb/rag1: 10-45), absence (cmyb/rag1: 0-10) foci. Quantifications are shown in (q), 3 independent experiments. r Number of coro1a:EGFP+/lyz:DsRed- cells entering thymus every 30 min in WT, atf7ip−/−, bach2b MO-injected atf7ip−/− embryos, n = 3. s, t Confocal imaging showing thymic lymphoid progenitors (white circles) of WT, atf7ip−/− and bach2b MO-injected atf7ip−/− embryos on Tg(coro1a:EGFP/lyz:DsRed) background at 72 hpf. Quantification is shown in (t), n = 3. Scale bar, 100 μm (p, s). Quantifications (f–h, l–o, q, r, t) are represented as mean ± SD; ns, P > 0.05, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Student’s t test (unpaired, two-tailed). Source data are provided as a Source Data file.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Nat. Commun.