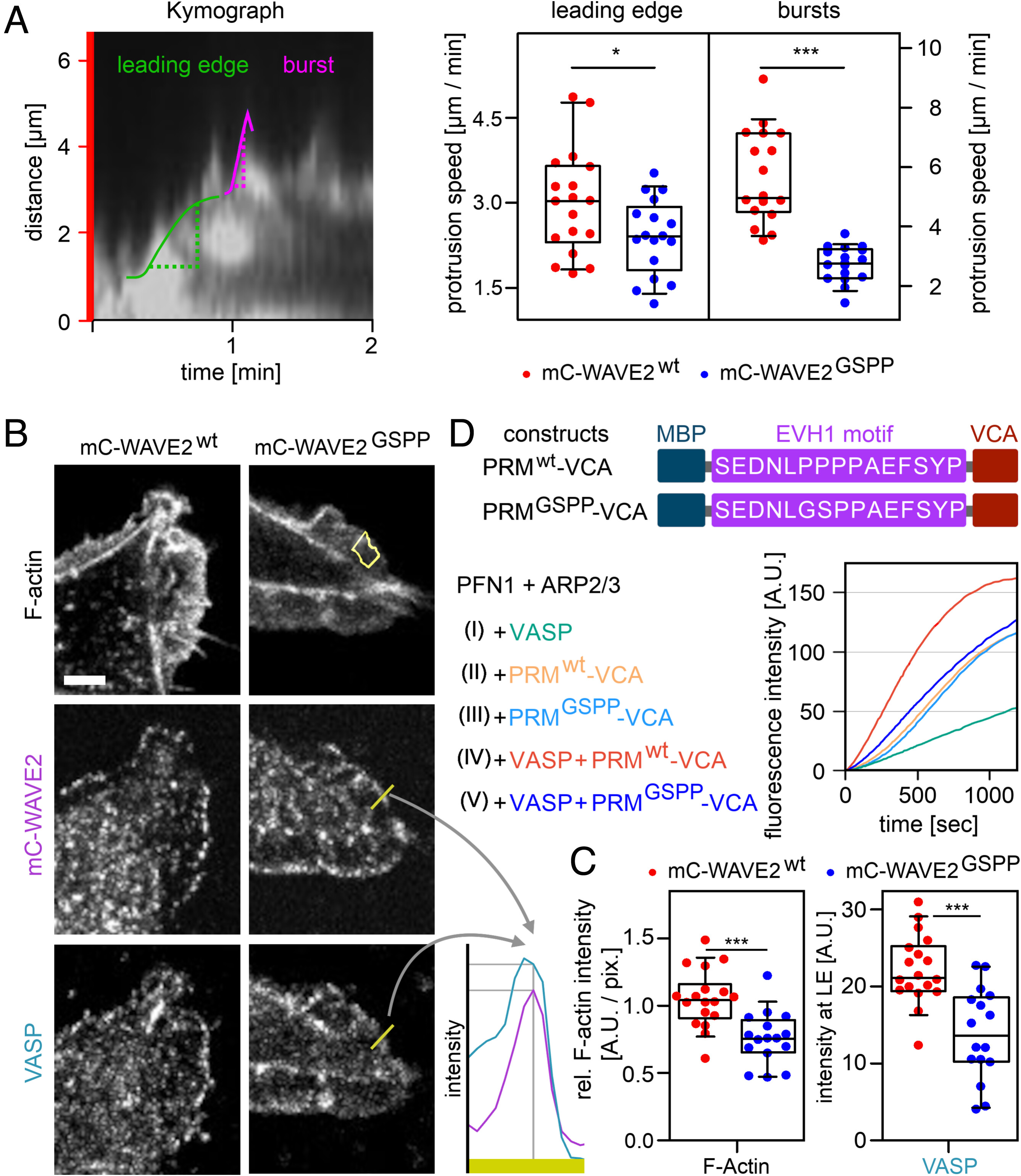
Fig. 3 EVH1–WAVE2 interaction modulates actin polymerization rates and lamellipodial protrusion velocity. (A) Lamellipodial protrusions. Left panel: A kymograph of membrane protrusions upon stimulation with EGF after starvation in MDA-MB-231 cells with time on the x-Axis and lamellipodial protrusion along a red bar (SI Appendix, Fig. S6) from TIRF micrographs. The protrusion velocity of the leading edge (green) and fast membrane burst protrusion (pink) were calculated. Right panel: Statistical evaluation of protrusion speeds for mC-WAVE2wt and mC-WAVE2GSPP cells. Data from three independent experiments are shown. T test analysis for leading edge (*P ≤ 0.05) and Mann-Whitney test for the bursts (***P ≤ 0.001). (B) Immunofluorescence stains (confocal micrographs) for F-actin, mCherry-WAVE2 constructs, and VASP in MDA-MB-231 cells stimulated with EGF after starvation and the statistical evaluation (C). Lamellipodia were identified via F-actin stain, and signal intensity was measured (the yellow area in the Upper Right panel represents the measured region of interest). The profile of WAVE2 and VASP (yellow bar in Middle and Lower Right panels) was plotted (Lower Right panel) and the intensities for both proteins were measured at maximal WAVE2 signal, indicating the leading edge. The scale bar (white) represents 5 µm. (C) Statistical evaluation of immunofluorescence stains. The F-actin intensity in the lamellipodia (Left panel) and VASP (Right panel) in the leading edge was evaluated. Data from two independent experiments. T test analysis (***P ≤ 0.001)). (D) In vitro actin polymerization assay of pyrene-labeled G-actin in the presence of profilin (PFN1) and ARP2/3 complex. WAVE2 constructs: maltose binding protein (MBP); WAVE2 motif recognized by EVH1 (wt) and the nonbinding mutant (GSPP); VCA domain. Raw fluorescence signal over time.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Proc. Natl. Acad. Sci. USA