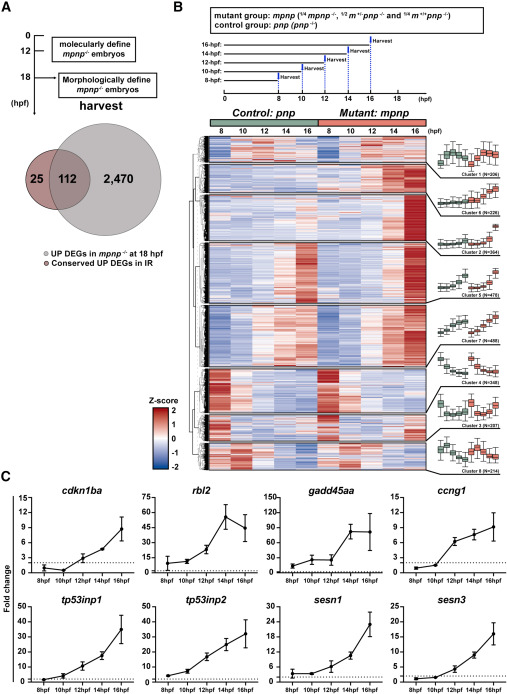
Fig. 6 Comparing 137 conserved p53 dependent IR induced genes with DEGs in mpnp−/− datasets (A) Venn graph displaying the overlapping genes between 137 conserved UP DEGs in both zebrafish and mouse with IR-irradiation and 2,582 upregulated (UP) DEGs in mpnp−/− versus sibling controls at 18 hpf. Experimental timeline showing the timepoint that distinguish mpnp−/− embryos from sibling controls and how to harvest RNA samples at the time (Top panel). (B) Heatmap showing transcriptional changes for 2,582 DEGs over time in early mpnp−/− datasets (8, 10, 12, 14, and 16 hpf). Each timepoint was measured in duplicate, and the averages of the duplicates were used for each group. Z-scores, calculated from TPM values calculated across all samples, were used to standardize gene expression levels before clustering. The experimental workflow for sample collection is shown in the top panel. Note that mutant samples at each timepoint were diluted 4-fold by their sibling controls (¼ mpnp−/− and ¾ sibling controls). Genes with similar expression patterns over time were grouped into eight clusters, with their trends and cluster sizes shown in the right panel. (C) Line graphs showing the kinetics of 8 out of 24 GOIs in early mpnp datasets, with the dashed line indicating a fold change of 2. Expected counts calculated with RSEM were used for creating line graphs. Line graphs for the remaining 16 GOIs are shown in Figure S8B.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ iScience