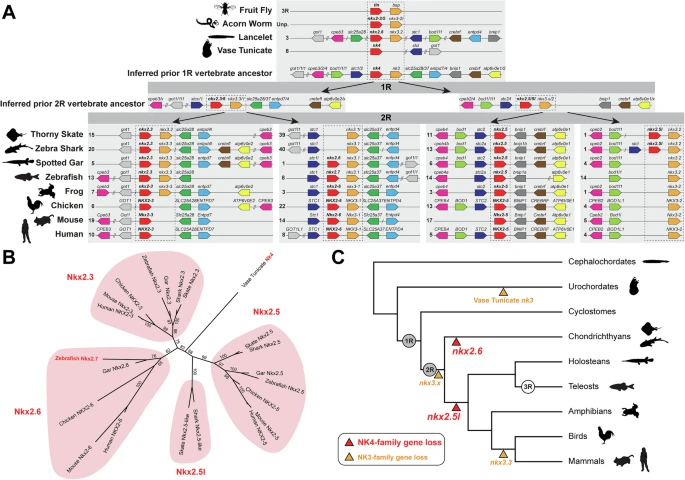
Fig. 1 Evolution of NK4-family genes in bilaterian animals.A Conserved microsynteny of NK4-family genes (nkx2.3, nkx2.6/7, nkx2.5, nkx2.5l) with neighboring genes from NK3 family (nkx3.1, nkx3.2, nkx3.3) and other families (cpeb2/3/4, got1/1l1, bod1/1l1, stc1/2, nkx3.1/2/3, slc25a28/37, entpd4/7, bnip1, crebrf/rfl, atp6v0e1/2). Single slashes indicate the presence of a single other gene, while a double slash indicates the presence of two or more other genes. One ancestral chromosomal locus with inferred genes was present in the vertebrate ancestor, prior to the first round (1 R) of vertebrate whole genome duplication (WGD) and two ancestral chromosomal loci with inferred genes are shown prior to the second round (2 R) of WGD. Four chromosomal loci were identified for extant vertebrate NK4-family genes: nkx2.3, nkx2.6/7 (not found in thorny skate and zebra shark), nkx2.5, and nkx2.5l (not found in investigated bony fishes). Zebrafish retained only one copy of each NK4- and NK3-family gene following 3 R, and zebrafish nkx2.7 shares the conserved synteny with vertebrate Nkx2.6. B The phylogenetic reconstruction of NK4-family genes places the tunicate Nk4 gene at the base of two nodes, one node leading to nkx2.3 and nkx2.6 subfamilies and another to nkx2.5 and nkx2.5l subfamilies. Zebrafish nkx2.7 and gar nkx2.6 cluster with the amniote Nkx2-6 genes. C Summary of whole genome duplications and NK4- and NK3-family gene losses in bilaterian animals.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Nat. Commun.