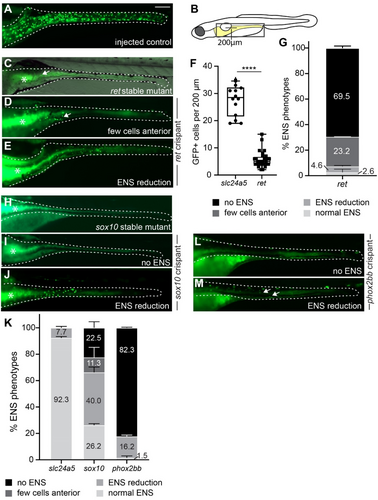
Fig. 1 F0 crispants targeting ret, sox10, or phox2bb recapitulate known ENS phenotypes. (A–E) F0 crispants targeting ret phenocopy the lack of phox2bb:GFP+ ENS cells (green) in stable ret mutant larvae (C) in comparison to wildtype-level ENS cells in control larvae (A). Compare ENS phenotypes between (C) and (D), arrows point to remaining ENS neurons in the anterior gut. (E) A small portion of ret F0 crispants show ENS cell reduction (B) Schematic of zebrafish larvae, the gray boxed area indicates the area shown in A-M. Line indicates length used for quantification of anterior neuron numbers in ret crispants in (F). (F) Significantly fewer GFP+ cells are present within 200 um of anterior gut in ret F0 crispants compared to control injected larvae (p < 0.0001, 2 experiments, % shown as mean ± SEM, each data point equals one larva). (G) % of ENS phenotypes in ret F0 crispants (2 experiments, % shown as mean ± SEM). (H–J) F0 crispants targeting sox10 (I) phenocopy the lack of GFP+ ENS cells in stable sox10 mutant larvae (H). A small portion of F0 crispants targeting sox10 show ENS cell reduction (J). (K) % of ENS phenotypes in control slc24a5, sox10, and phox2bb F0 crispants (≥ 2 experiments, % shown as mean ± SEM). (L, M) The majority of F0 crispants lack GFP+ ENS cells (L). A small portion have reduction in ENS cells (M). (A, C–E, H–J, L–M) Whole-mount side views at 6 days post fertilization (dpf). Asterisks: autofluorescence in gut epithelium; dashed line: Gut outline. Scale bar = 100 μm.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Neurogastroenterol. Motil.