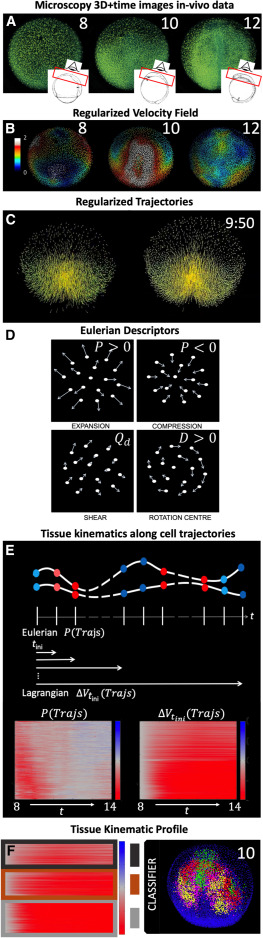
Fig. 1 Construction of the biomechanical map from the reconstructed cell lineage (A–C) WT specimen wt1 imaged live between 6 and 12 hpf, animal pole (AP) view, rendered in 3D using the Mov-IT software. Scale bar 50 . (A) Nuclear staining (green) shown at 8, 10 and 12 hpf as indicated in each panel. The insets depict the imaged volume and field of view. (B) Cell velocity magnitudes obtained from the cells’ regularized displacement field (see STAR Methods, Figure S2) and represented in as indicated by color bar. (C) Original cell trajectories (left panel) vs. regularized ones (right panel). Cell nuclei are shown as cubes and 5-time-step segments of their trajectories are shown by lines. (D) Schematics of simple cell flows, associated tissue deformation patterns, and corresponding IDG descriptors: expansion ( ), compression (), shear (), and rotation ( ). (E) Schematic illustration depicting the computation of tissue kinematics descriptors along cell trajectories. Tissue deformation invariants are computed at each point along the cell trajectories from the instantaneous (Eulerian) or finite time (Lagrangian) deformation gradients (e.g., and , respectively, where red indicates compression and blue indicates expansion blue). The resulting time-dependent profiles are plotted for a selection of cells between 8 ( ) and 14 hpf, leading to an atlas of tissue kinematics maps along cell trajectories. (F) Overview of the two-step clustering approach used to identify embryonic domains undergoing distinct tissue kinematics. First, we cluster each map within the atlas to identify representative tissue kinematics profiles (RTKPs, dark gray, brown, light gray). Second, the RTKPs are used to classify cells within the embryo, revealing connected embryonic domains that experience distinct spatiotemporal evolution of tissue kinematics (red, magenta, green and yellow).
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ iScience