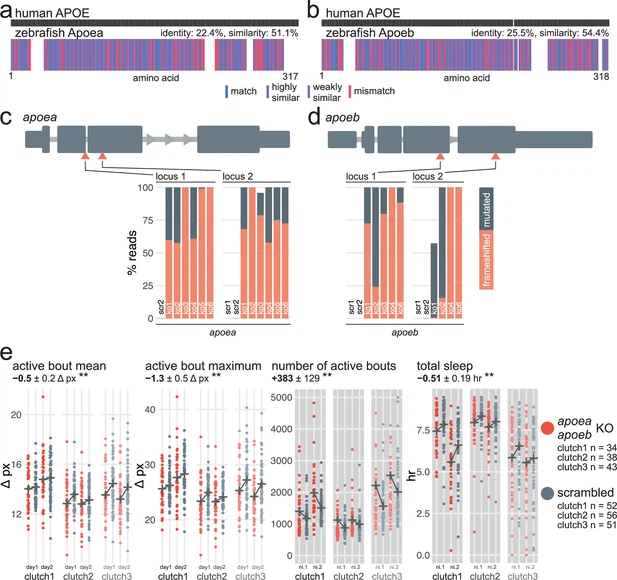
Fig. 4 - Supplemental 1 apoea/apoeb double F0 knockouts have subdued swimming bouts during the day and sleep less at night. (a) Human APOE amino acid sequence (top) aligned to zebrafish Apoea amino acid sequence (bottom). In the zebrafish protein, each amino acid (vertical bar) is coloured based on its similarity with the human protein. White gaps are added when additional residues are present in the other sequence. (b) Human APOE amino acid sequence (top) aligned to zebrafish Apoeb amino acid sequence (bottom), as in (a). (c) (top) Schematic of apoea in the 5′–3′ genome direction. Exons are in dark grey; tall exons are protein-coding, small are 5′- or 3′-UTR. Light grey lines are introns, and grey arrows represent the direction of transcription. Orange arrowheads mark the target loci. Exons and introns are on different scales. (bottom) Percentage of reads mutated (height of each bar, with orange representing percentage with a frameshift mutation) at each targeted locus of apoea. scr, scrambled-injected control larva; ko, apoea F0 knockout larva. Across F0 knockout samples: 99.6 ± 1.2% mutated reads, 77.5 ± 17.8% of all reads had a frameshift mutation. (d) (top) Schematic of apoeb, as in (c). (bottom) Percentage of reads mutated (height of each bar, with orange representing percentage with a frameshift mutation) at each targeted locus of apoeb. scr, scrambled-injected control larva; ko, apoeb F0 knockout larva. Across F0 knockout samples: 95.2 ± 14.2% mutated reads, 64.6 ± 39.9% of all reads had a frameshift mutation. In (c) and (d), the numbers refer to individual animals. For example, ko6 refers to an individual apoea/apoeb double F0 knockout larva for which mutations at the four targeted loci are plotted. (e) Parameter plots for three clutches of apoea/apoeb double F0 knockout larvae and scrambled-injected siblings. Each dot represents one larva during one day (left) or night (right). Black crosses mark the group means. (left) During the day, apoea/apoeb double F0 knockout larvae had more subdued swimming bouts than scrambled-injected siblings (active bout mean, ** p=0.006; active bout maximum, ** p=0.004). (right) At night, apoea/apoeb double F0 knockout larvae were performing more swimming bouts (number of active bouts, ** p=0.003) and sleeping less (total sleep, ** p=0.009) than scrambled-injected siblings. Six datapoints from four apoea/apoeb F0 knockout larvae fall outside the Y axis limit of the number of active bouts (7229–13,975 bouts). Statistics by likelihood-ratio test on linear mixed effects models. ni., night.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Elife