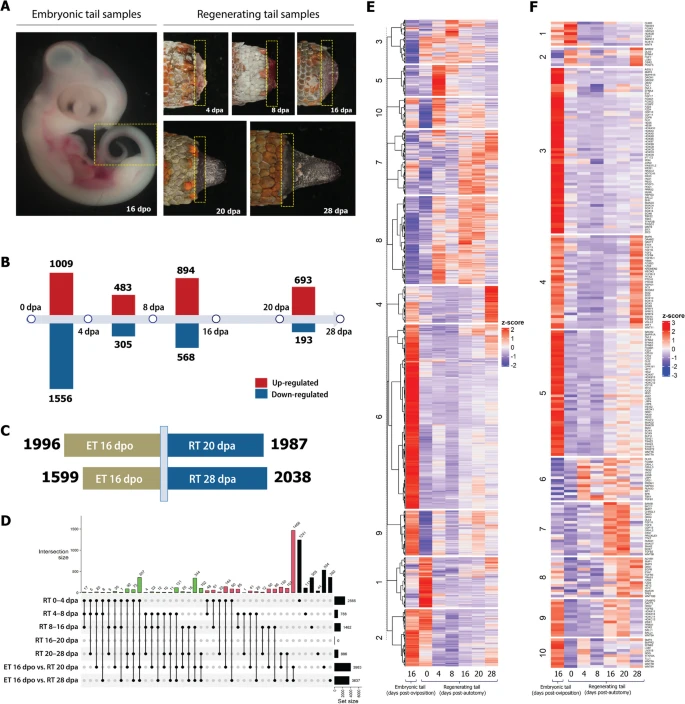
Fig. 1 Transcriptome analysis shows a distinctive gene expression profile at each stage of regenerating tail and embryonic tail. A Gecko samples used for transcriptome analysis (indicated by boxed areas). Note that we also included adult tail stump/0 dpa for transcriptome analysis (figure not shown). B Number of up- and down-regulated genes during tail regeneration stages. Differential gene expression analysis by stage-wise comparison. Three biological replicates each stage. Significant differential expression (DE) genes with adjusted p-value < 0.01 and L2FC ≥ 1. Statistical test results can be found in Additional file 6: Suppl_ 1. C Number of genes regulated in regenerating tail vs. embryonic tail. ET embryonic tail, RT regenerating tail. Three biological replicates each stage. Significant differential expression (DE) genes with adjusted p-value < 0.01 and L2FC ≥ 1. D UpSet of cross-state showing the comparison between different stages of regeneration and tail development. ET embryonic tail, RT regenerating tail. E Heatmap of 6817 significant differentially expressed genes (whole transcriptome) in embryonic and regenerating tails of the Tokay Gecko. F Heatmap of 236 significant differentially expressed genes (toolkit genes only) in embryonic and regenerating tails of the tokay gecko
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ BMC Biol.