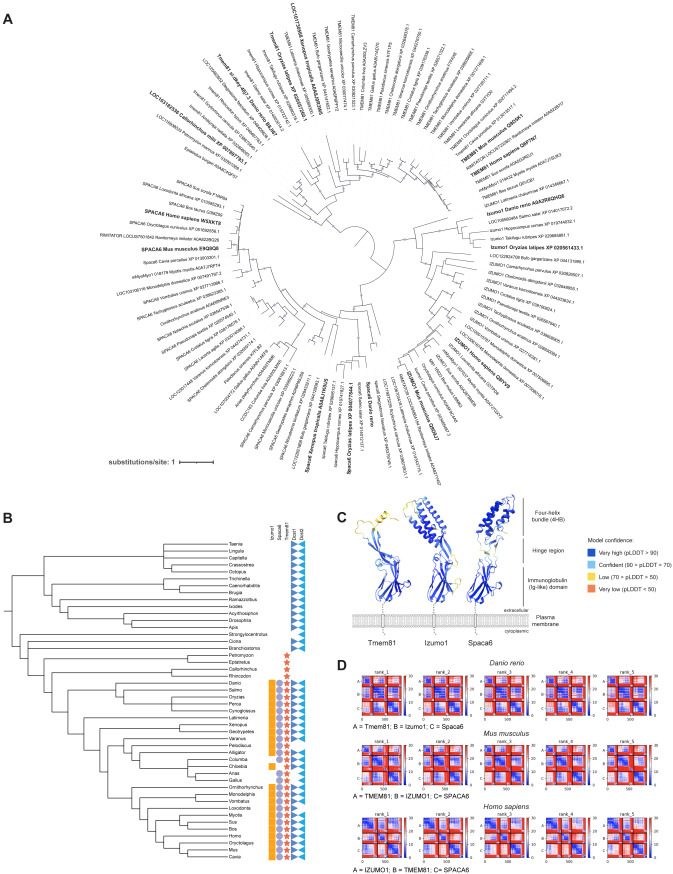
Fig. s1 Conservation of vertebrate fertility factors and predicted interaction between Izumo1, Spaca6, and Tmem81, related to Figure 1 (A) Phylogenetic tree of Izumo1, Spaca6, and Tmem81 proteins. Bootstrap values > 94 are indicated by filled purple circles. Branch lengths represent the inferred number of amino acid substitutions per site, and branch labels are composed of gene name (if available), genus, species, and accession number. (B) Taxonomic tree of Izumo1 (orange square), Spaca6 (blue circle), Tmem81 (red star), Dcst1 (dark blue triangle), and Dcst2 (light blue triangle) across vertebrates and invertebrates. (C) Tertiary structure predictions of zebrafish Tmem81, Izumo1, and Spaca6. Each amino acid is shaded as indicated according to its predicted local distance difference test (pLDDT) score as a confidence measure. Predictions were generated by AlphaFold2.36 (D) PAE plots of the top 5 models of the predicted trimeric complexes in zebrafish (top), mouse (center), and human (bottom). Units: amino acid residues; color bar: expected position error in angstroms.
Reprinted from Cell, 187(25), Deneke, V.E., Blaha, A., Lu, Y., Suwita, J.P., Draper, J.M., Phan, C.S., Panser, K., Schleiffer, A., Jacob, L., Humer, T., Stejskal, K., Krssakova, G., Roitinger, E., Handler, D., Kamoshita, M., Vance, T.D.R., Wang, X., Surm, J.M., Moran, Y., Lee, J.E., Ikawa, M., Pauli, A., A conserved fertilization complex bridges sperm and egg in vertebrates, 7066-7078.e22, Copyright (2024) with permission from Elsevier. Full text @ Cell