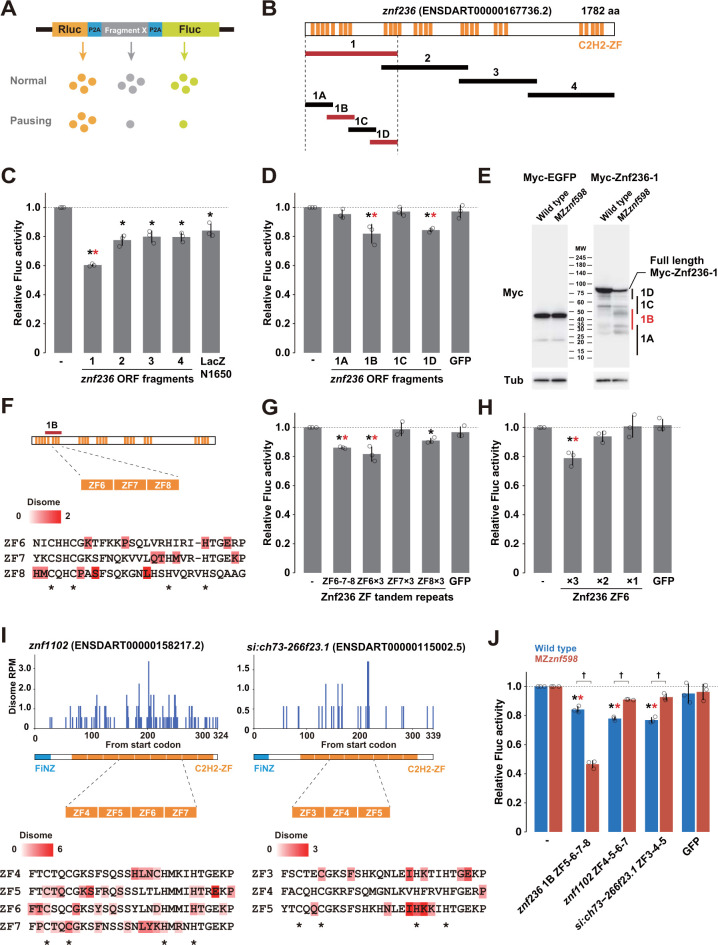
Fig 5 Analysis of ribosome stalling sequences in C2H2-ZF mRNAs.
(A) A schematic of the tandem ORF assay. The Rluc ORF (orange) and Fluc ORF (light green) are separated by 2 P2A translation skipping sequences (blue). The sequence to be tested (shown as X in gray) is inserted between the 2 P2A sequences. (B) Schematic of the