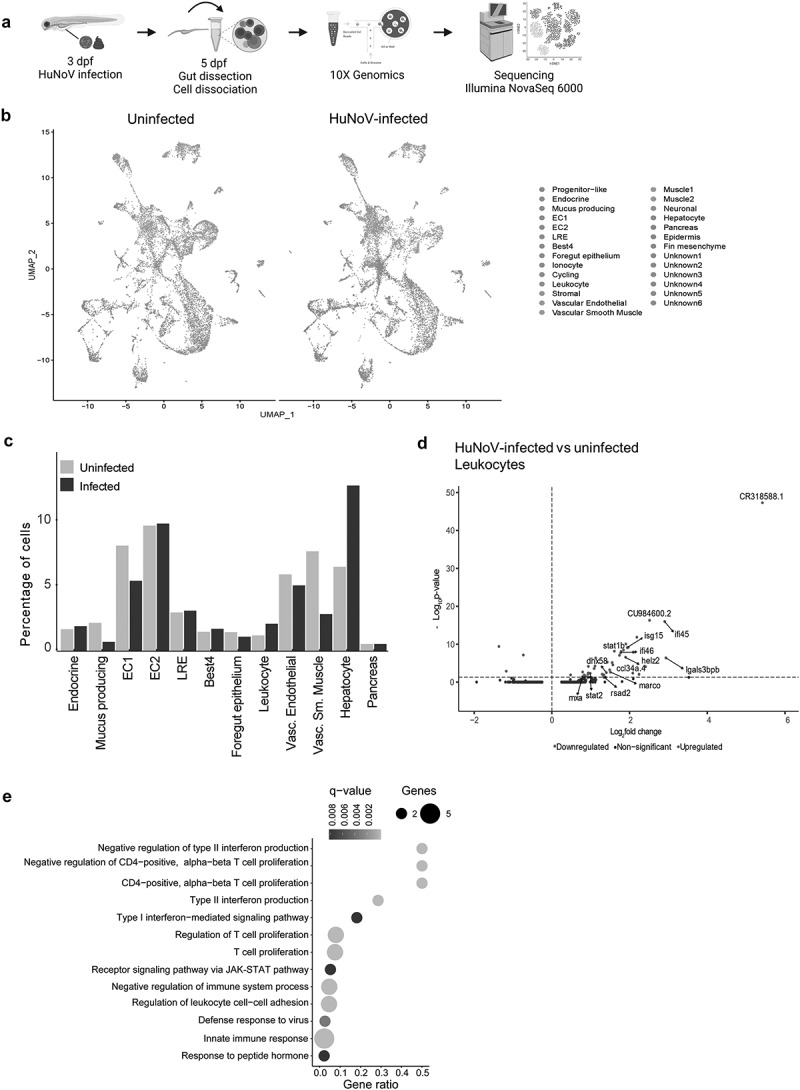
Figure 2.
A scRNA-seq analysis on dissected zebrafish larval intestines shows an increase in leukocytes and an important role in the antiviral immune response. (a) Experimental design for transcriptional profiling of single cells in the zebrafish intestine. (b) Aggregated data of all intestinal cells (