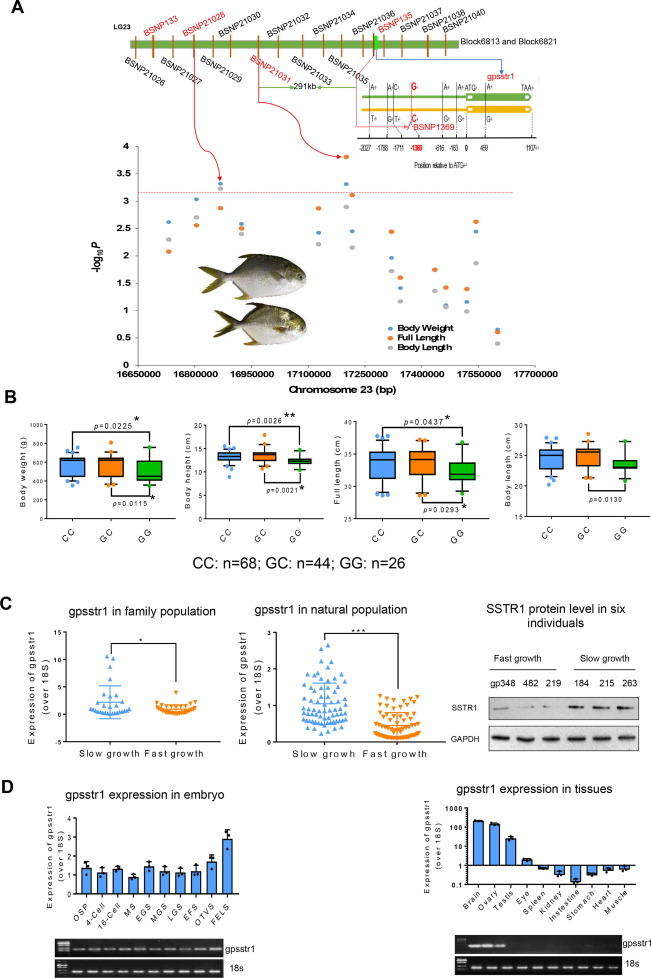
Fig. 6 QTL fine mapping and identification of gene closely associated growth. A. Fine mapping of candidate gene gpsstr1. According to the confirmed SNPs (BSNP132, BSNP135), underlying bin markers of Block6813 and Block6821, a total of 14 SNPs were developed to genotype a panel of 146 natural-collected individuals by KASP Assays. Manhattan plot of haplotype-based regional association analysis was displayed. The red dotted line represents the significance threshold (P = 0.01/14) (bottom left corner). The SNP (-1369) in the promoter region of gpsstr1 was verified by Sanger sequencing. B. The natural growing group were grouped according to their genotypes (CC, GC&GG). Two-tailed Student's t-test was used to calculate the P value between different groups (bottom right corner). For all the box charts: minimum value = lower whisker, maximum value = upper whisker, median = middle value of the box, lower quartile = median of the lower half of dataset, upper quartile = median of the upper half of dataset, datapoint outside of whiskers = potential outlier. C. Expression of gpsstr1 in 64 family population individuals (left) and 146 natural population individuals (middle) of golden pompano, respectively, using qPCR method. According to their growth trait data, golden pompano fishes were classified into slow and fast-growth groups. The protein levels of SSTR1 in three fast-growth (numbered gp238, gp482 and gp219) and three slow-growth individuals (numbered gp184, gp215 and gp263) were measured by Western blotting method. Band intensity represents protein level. The data represent as mean ± sem. *p < 0.05, **p < 0.01, ***p < 0.001 represents statistically significant. D. Expression profiles of gpsstr1 in 10 embryo stages and 10 tissues of golden pompano by using qPCR (up) and reverse transcription PCR (electrophoretic bands, down). 18S RNA was used as a reference. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ J Adv Res