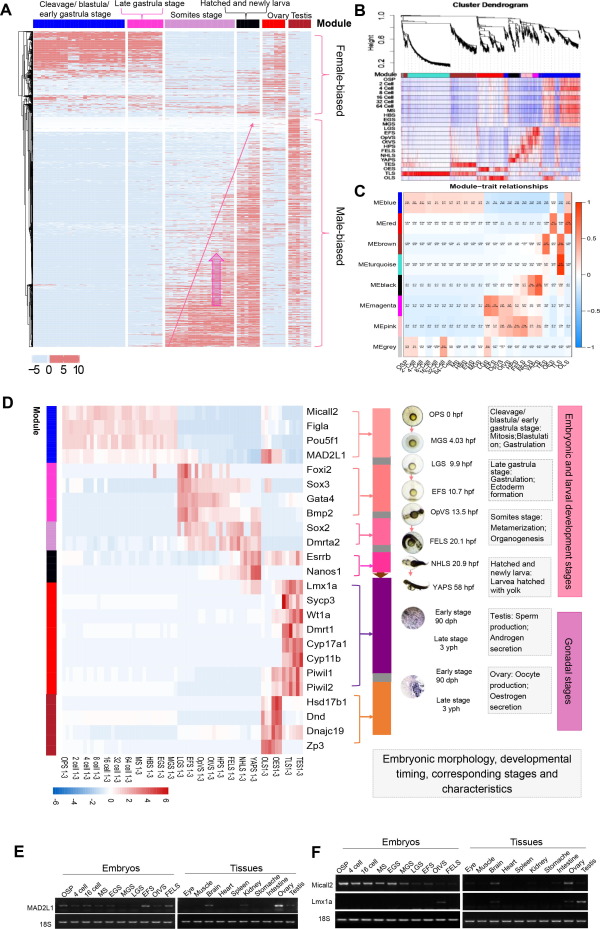
Fig. 5 Sex-biased gene expression throughout development. A. Heatmap of the sex-related genes in 19 embryonic developmental stages were clustered based on the FPKM value. B. Heatmap of the gene co-expression network. Clustering dendrograms of all sex-related genes, with dissimilarity based on the topological overlap and assigned module colors. Seven modules were clustered and displayed in different colors. C. Module-trait associations. Each column corresponds to a development stage, and each row corresponds to a module. The number in the rectangle is the correlation coefficient, and the number in brackets is the corresponding p value. D. Heatmap of the selected 24 sex-related stage-specific genes filtered by previously documented and the KME value > 0.7. In each module, the corresponding development stage and time were listed in the right part. E F. PCR validation of MAD2L1 (E), Micall2 and Lmx1a (F) in embryos and organs of golden pompano. 18S RNA was used as a reference. OSP, oosperm; MS, morula stage; EGS, early gastrula stage; MGS, middle gastrula stage; LGS, late gastrula stage; EFS, embryo formed stage; OTVS, otocyst vesicle stage; FELS, formation of the eye lens.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ J Adv Res