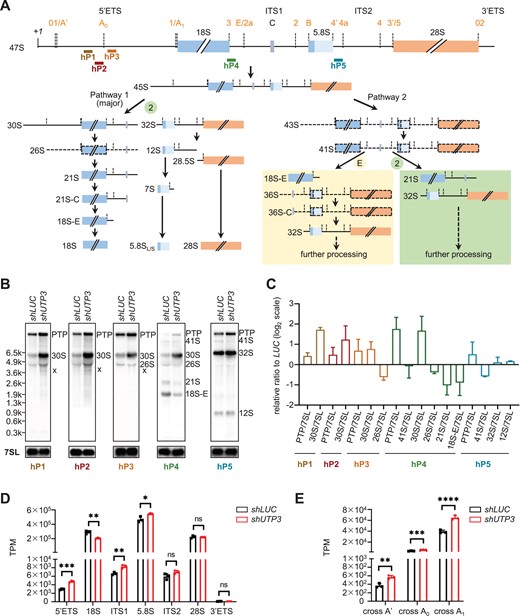
Fig. 9 Depletion of human UTP3 mainly impairs the 5′ETS processing at the A0-site. (A) Diagram showing the human rDNA genomic structure and positions of the five probes (hP1 to hP5) for Northern blot. Short-lived precursors are represented with dotted lines. (B) Comparison of the processed rRNA species in shLUC and shUTP3 cells by Northern blot using three 5′ETS (hP1, hP2, hP3) probes, one ITS1 (hP4) probe and one ITS2 (hP5) probe. The intermediate products of pre-rRNA processing in cells were highlighted according to (A). PTP: primary transcript plus, co-migrating 47S and 45S pre-rRNAs; x: aberrant band. (C) Ratio analysis of multiple precursors (RAMP) derived from three independent Northern blot experiments. Different colors of bars represent analyses derived from distinct probes as indicated. (D) Graph showing the statistical analysis of the TPM-based sequence reads derived from the total RNA-seq data for 5′ETS, 18S, ITS1, 5.8S, ITS2, 28S and 3′ETS in shLUC and shUTP3 cells. (E) Statistical analysis of reads encompassing A′, A0 and A1 cleavage sites in shLUC and shUTP3 cells.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Nucleic Acids Res.