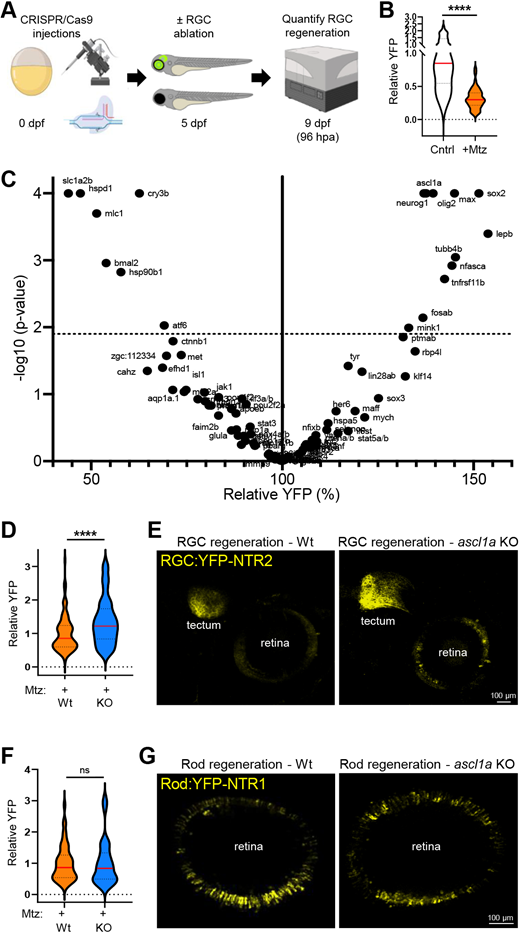
Fig. 4 CRISPR screen identifies genes regulating RGC regeneration. (A) Experimental setup for CRISPR/Cas9-based ‘crispant’ knockout (KO) screen for genes that regulate RGC regeneration. (B) Plate reader-based quantification of RGC regeneration at 96 hpa. Relative YFP levels in RGC ablated larvae (+Mtz) are normalized to unablated controls (Cntr). (C) Volcano plot of RGC regeneration (relative YFP levels) for all Mtz-treated RGC:YFP-NTR2 crispant KO larvae (dashed line indicates adjusted P-value cutoff of 0.01; minimum sample size n=8, median sample size n=16). (D) Plate reader-based quantitative comparison of RGC regeneration levels (YFP) between Mtz-treated control (Wt) and ascl1a crispant (KO) larvae at 96 hpa. (E) Representative confocal images of RGC regeneration in Mtz-treated control (Wt) and ascl1a crispant (KO) larvae at 168 hpa. (F) Plate reader-based quantitative comparison of rod cell regeneration levels (YFP) between Mtz-treated control (Wt) and ascl1a crispant (KO) larvae at 96 hpa. (G) Representative confocal images of rod cell regeneration in Mtz-treated control (Wt) and ascl1a crispant (KO) larvae at 168 hpa. ****P≤0.0001; ns, not statistically significant.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Development