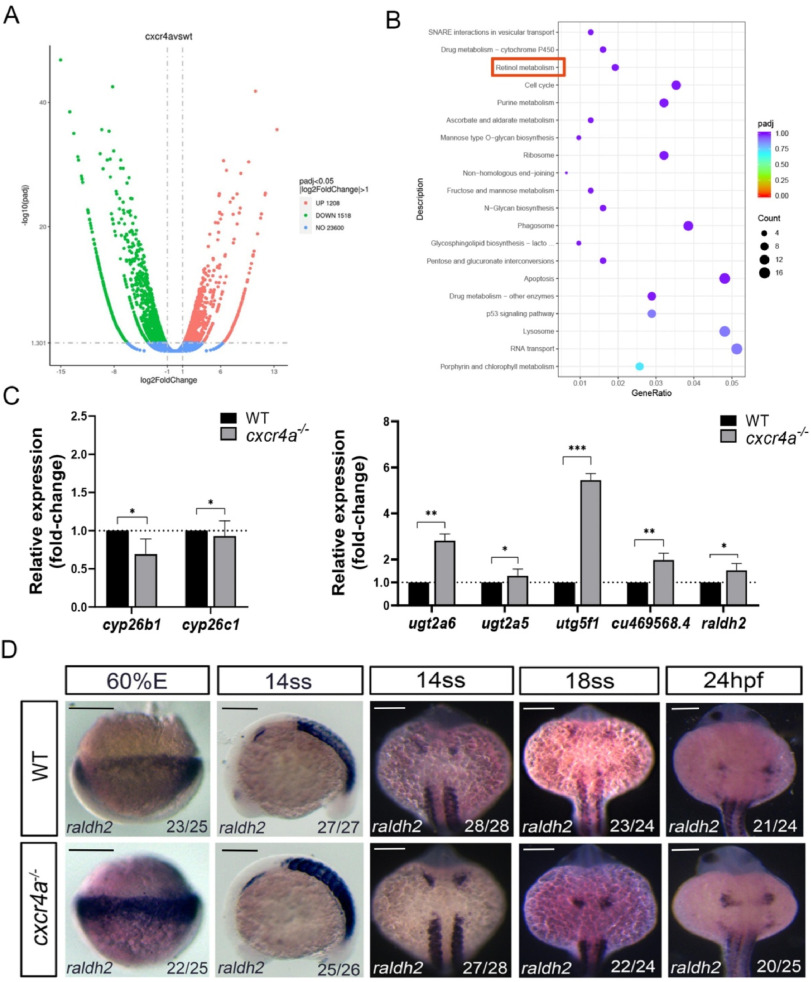
Fig. 2 RA signaling pathway is up-regulated in cxcr4a mutants (A) Volcano map of genes expressed differently between control embryos and cxcr4a mutant embryos. The up-regulated genes are indicated by red dots and down-regulated genes by green dots. (B) KEGG rich distribution map. The retinol metabolism pathway is also disturbed, which is marked by red box. (C) The genes being involved in RA signaling are evaluated using qRT-PCR. In cxcr4a mutants cyp26b1 and cyp26c1 are down-regulated and ugt2a5, ugt2a6, ugt5f1, cu469568.4 and raldh2 were all up-regulated. (D) The expression of raldh2 is examined using in situ hybridization at 60 % epiboly, 14SS, 18SS and 24hpf, it is up-regulated at all stages in cxcr4a mutants. Error bars indicate standard deviation of the mean. Bar, 50 μm *P<0.05; **P<0.01; ***P<0.001. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Biochem Biophys Rep