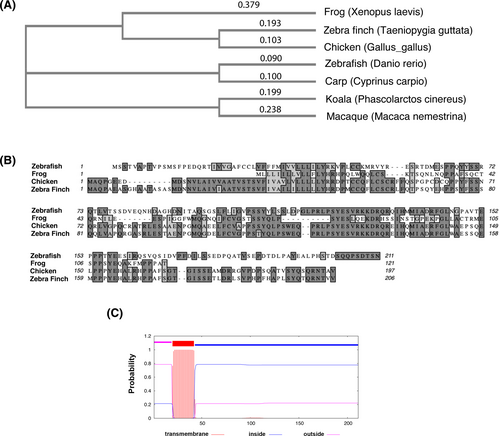
Fig. 1 Drish conservation between different species and structure prediction. (A) The phylogenetic tree analysis of Drish proteins from different species. The cladogram of Neighbor-joining tree without distance corrections was generated using Clustal Omega multiple sequence alignment tool at EMBL-EBI.68 (B) Alignment between amino acid sequences of the Danio rerio Drish and amino acids from three other species is shown. The dark gray highlight shows identical amino acid residues, while light gray highlight shows similar amino acid residues between different species. The amino acid sequence alignment was performed using MacVector 18.0 software. The following sequences were used for alignments in (A, B): Cyprinus carpio XP_042576826; Gallus gallus XP_429916.4; Macaca nemestrina XP_011723858.1; Phascolarctos cinereus XP_020828228.1; Taeniopygia guttata XP_030143607.1; Xenopus laevis XELAEV_18044022mg; Danio rerio XP_009298140.1. (C) The transmembrane helices were analyzed by TMHMM2.0 software.35 The graphical output shows the posterior probabilities for outside (luminal, aa 1 to aa 21), transmembrane (aa 22 to aa 42), and inside (cytoplasmic, aa 43 to aa 211) regions.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Dev. Dyn.